sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans
- PMID: 14534247
- PMCID: PMC6740817
- DOI: 10.1523/JNEUROSCI.23-27-09133.2003
sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans
Abstract
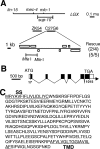
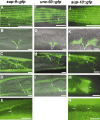
Genetic studies of sup-9, unc-93, and sup-10 strongly suggest that these genes encode components of a multi-subunit protein complex that coordinates muscle contraction in Caenorhabditis elegans. We cloned sup-9 and sup-10 and found that they encode a two-pore K+ channel and a novel transmembrane protein, respectively. We also found that UNC-93 and SUP-10 colocalize with SUP-9 within muscle cells, and that UNC-93 is a member of a novel multigene family that is conserved among C. elegans, Drosophila, and humans. Our results indicate that SUP-9 and perhaps other two-pore K+ channels function as multiprotein complexes, and that UNC-93 and SUP-10 likely define new classes of ion channel regulatory proteins.
Figures






Similar articles
-
The Caenorhabditis elegans iodotyrosine deiodinase ortholog SUP-18 functions through a conserved channel SC-box to regulate the muscle two-pore domain potassium channel SUP-9.PLoS Genet. 2014 Feb 20;10(2):e1004175. doi: 10.1371/journal.pgen.1004175. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24586202 Free PMC article.
-
The Caenorhabditis elegans unc-93 gene encodes a putative transmembrane protein that regulates muscle contraction.J Cell Biol. 1992 Apr;117(1):143-55. doi: 10.1083/jcb.117.1.143. J Cell Biol. 1992. PMID: 1313436 Free PMC article.
-
An abnormal ketamine response in mutants defective in the ryanodine receptor gene ryr-1 (unc-68) of Caenorhabditis elegans.J Mol Biol. 1997 Apr 11;267(4):849-64. doi: 10.1006/jmbi.1997.0910. J Mol Biol. 1997. PMID: 9135117
-
An evolutionarily conserved family of accessory subunits of K+ channels.Cell Biochem Biophys. 2006;46(1):91-9. doi: 10.1385/CBB:46:1:91. Cell Biochem Biophys. 2006. PMID: 16943626 Review.
-
Potassium channels in C. elegans.WormBook. 2005 Dec 30:1-15. doi: 10.1895/wormbook.1.42.1. WormBook. 2005. PMID: 18050399 Free PMC article. Review.
Cited by
-
Complex Locomotion Behavior Changes Are Induced in Caenorhabditis elegans by the Lack of the Regulatory Leak K+ Channel TWK-7.Genetics. 2016 Oct;204(2):683-701. doi: 10.1534/genetics.116.188896. Epub 2016 Aug 17. Genetics. 2016. PMID: 27535928 Free PMC article.
-
Differential modulation of C. elegans motor behavior by NALCN and two-pore domain potassium channels.PLoS Genet. 2022 Apr 28;18(4):e1010126. doi: 10.1371/journal.pgen.1010126. eCollection 2022 Apr. PLoS Genet. 2022. PMID: 35482723 Free PMC article.
-
UNC93B1 interacts with the calcium sensor STIM1 for efficient antigen cross-presentation in dendritic cells.Nat Commun. 2017 Nov 21;8(1):1640. doi: 10.1038/s41467-017-01601-5. Nat Commun. 2017. PMID: 29158474 Free PMC article.
-
Genome-wide view of cell fate specification: ladybird acts at multiple levels during diversification of muscle and heart precursors.Genes Dev. 2007 Dec 1;21(23):3163-80. doi: 10.1101/gad.437307. Genes Dev. 2007. PMID: 18056427 Free PMC article.
-
Natural Variation in plep-1 Causes Male-Male Copulatory Behavior in C. elegans.Curr Biol. 2015 Oct 19;25(20):2730-7. doi: 10.1016/j.cub.2015.09.019. Epub 2015 Oct 8. Curr Biol. 2015. PMID: 26455306 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
