Copper response element and Crr1-dependent Ni(2+)-responsive promoter for induced, reversible gene expression in Chlamydomonas reinhardtii
- PMID: 14555481
- PMCID: PMC219375
- DOI: 10.1128/EC.2.5.995-1002.2003
Copper response element and Crr1-dependent Ni(2+)-responsive promoter for induced, reversible gene expression in Chlamydomonas reinhardtii
Abstract
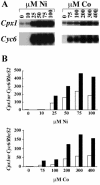
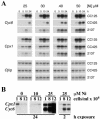
The Cpx1 and Cyc6 genes of Chlamydomonas reinhardtii are activated in copper-deficient cells via a signal transduction pathway that requires copper response elements (CuREs) and a copper response regulator defined by the CRR1 locus. The two genes can also be activated by provision of nickel or cobalt ions in the medium. The response to nickel ions requires at least one CuRE and also CRR1 function, suggesting that nickel interferes with a component in the nutritional copper signal transduction pathway. Nickel does not act by preventing copper uptake/utilization because (i) holoplastocyanin formation is unaffected in Ni(2+)-treated cells and (ii) provision of excess copper cannot reverse the Ni-dependent activation of the target genes. The CuRE is sufficient for conferring Ni-responsive expression to a reporter gene, which suggests that the system has practical application as a vehicle for inducible gene expression. The inducer can be removed either by replacing the medium or by chelating the inducer with excess EDTA, either of which treatments reverses the activation of the target genes.
Figures





References
-
- Akada, R., I. Hirosawa, M. Kawahata, H. Hoshida, and Y. Nishizawa. 2002. Sets of integrating plasmids and gene disruption cassettes containing improved counter-selection markers designed for repeated use in budding yeast. Yeast 19:393-402. - PubMed
-
- Atteia, A. 1994. Identification of mitochondrial respiratory proteins from the green alga Chlamydomonas reinhardtii. C. R. Acad. Sci. III. 317:11-19. - PubMed
-
- Bateman, J. M., and S. Purton. 2000. Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol. Gen. Genet. 263:404-410. - PubMed
-
- Bean, B., and A. Harris. 1979. Selective inhibition of flagellar activity in Chlamydomonas by nickel. J. Protozool. 26:235-240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

