Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development
- PMID: 14555692
- PMCID: PMC280558
- DOI: 10.1105/tpc.014043
Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development
Abstract
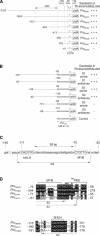
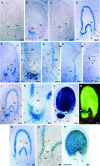
Anthocyanidin reductase encoded by the BANYULS (BAN) gene is the core enzyme in proanthocyanidin (PA) biosynthesis. Here, we analyzed the developmental mechanisms that regulate the spatiotemporal expression of BAN in the developing Arabidopsis seed coat. PA-accumulating cells were localized histochemically in the inner integument (seed body and micropyle) and pigment strand (chalaza). BAN promoter activity was detected specifically in these cells. Gain-of-function experiments showed that an 86-bp promoter fragment functioned as an enhancer specific for PA-accumulating cells. Mutations in regulatory genes of PA biosynthesis abolished BAN promoter activity (transparent testa2 [tt2], tt8, and transparent testa glabra1 [ttg1]), modified its spatial pattern (tt1 and tt16), or had no influence (ttg2), thus revealing complex regulatory interactions at several developmental levels. Genetic ablation of PA-accumulating cells targeted by the BAN promoter fused to BARNASE led to the formation of normal plants that produced viable yellow seeds. Importantly, these seeds had no obvious defects in endosperm and embryo development.
Figures







References
-
- Albert, S., Delseny, M., and Devic, M. (1997). BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 11, 289–299. - PubMed
-
- Bartel, B., and Matsuda, S.P.T. (2003). Seeing red. Science 299, 352–353. - PubMed
-
- Becraft, P.W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127, 4039–4048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

