Processive phosphorylation of alternative splicing factor/splicing factor 2
- PMID: 14555757
- PMCID: PMC240664
- DOI: 10.1073/pnas.1635129100
Processive phosphorylation of alternative splicing factor/splicing factor 2
Abstract
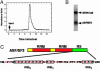
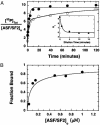
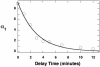
SR proteins, named for their multiple arginine/serine (RS) dipeptide repeats, are critical components of the spliceosome, influencing both constitutive and alternative splicing of pre-mRNA. SR protein function is regulated through phosphorylation of their RS domains by multiple kinases, including a family of evolutionarily conserved SR protein-specific kinases (SRPKs). The SRPK family of kinases is unique in that they are capable of phosphorylating repetitive RS domains with remarkable specificity and efficiency. Here, we carried out kinetic experiments specially developed to investigate how SRPK1 phosphorylates the model human SR protein, ASF/SF2. By using the start-trap strategy, we monitored the progress curve for ASF/SF2 phosphorylation in the absence and presence of an inhibitor peptide directed at the active site of SRPK1. ASF/SF2 modification is not altered when the inhibitor peptide (trap) is added with ATP (start). However, when the trap is added first and allowed to incubate for a specific delay time, the decrease in phosphate content of the enzyme-substrate complex follows a simple exponential decline corresponding to the release rate of SRPK1. These data demonstrate that SRPK1 phosphorylates a specific region within the RS domain of ASF/SF2 by using a fully processive catalytic mechanism, in which the splicing factor remains "locked" onto SRPK1 during RS domain modification.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

