MADS box genes control vernalization-induced flowering in cereals
- PMID: 14557548
- PMCID: PMC240751
- DOI: 10.1073/pnas.1635053100
MADS box genes control vernalization-induced flowering in cereals
Abstract
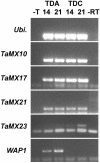
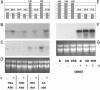
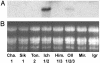
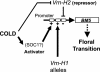
By comparing expression levels of MADS box transcription factor genes between near-isogenic winter and spring lines of bread wheat, Triticum aestivum, we have identified WAP1 as the probable candidate for the Vrn-1 gene, the major locus controlling the vernalization flowering response in wheat. WAP1 is strongly expressed in spring wheats and moderately expressed in semispring wheats, but is not expressed in winter wheat plants that have not been exposed to vernalization treatment. Vernalization promotes flowering in winter wheats and strongly induces expression of WAP1. WAP1 is located on chromosome 5 in wheat and, by synteny with other cereal genomes, is likely to be collocated with Vrn-1. These results in hexaploid bread wheat cultivars extend the conclusion made by Yan et al. [Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T. & Dubcovsky, J. (2003) Proc. Natl. Acad. Sci. USA 100, 6263-6268] in the diploid wheat progenitor Triticum monococcum that WAP1 (TmAP1) corresponds to the Vrn-1 gene. The barley homologue of WAP1, BM5, shows a similar pattern of expression to WAP1 and TmAP1. BM5 is not expressed in winter barleys that have not been vernalized, but as with WAP1, expression of BM5 is strongly induced by vernalization treatment. In spring barleys, the level of BM5 expression is determined by interactions between the Vrn-H1 locus and a second locus for spring habit, Vrn-H2. There is now evidence that AP1-like genes determine the time of flowering in a range of cereal and grass species.
Figures



References
-
- Law, C. N. (2000) Flowering Newslett. 29 22-25.
-
- Stelmakh, A. F. (1998) Euphytica 100 359-369.
-
- Snape, J. W., Butterworth, K., Whitechurch, E. & Worland, A. J. (2001) Euphytica 119 185-190.
-
- Pugsley, A. T. (1971) Aus. J. Agric. Res. 22 21-31.
-
- Halloran, G. M. (1975) Can. J. Genet. Cytol. 17 365-373.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

