Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency
- PMID: 14557628
- PMCID: PMC229253
- DOI: 10.1128/jvi.77.21.11425-11435.2003
Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency
Abstract
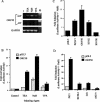
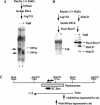
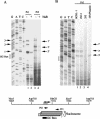
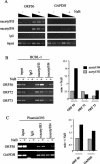
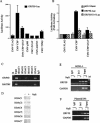
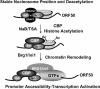
The switch from latent to lytic infection of Kaposi's sarcoma-associated herpesvirus is initiated by the immediate early transcriptional activator protein Rta/open reading frame 50 (ORF50). We examined the transcriptional regulation of the ORF50 core promoter in response to lytic cycle stimulation. We show that the ORF50 promoter is highly responsive to sodium butyrate (NaB) and trichostatin A (TSA), two chemicals known to inhibit histone deacetylases. The NaB and TSA responsive element was mapped to a 70-bp minimal promoter containing an essential GC box that binds Sp1/Sp3 in vitro and in vivo. Micrococcal nuclease mapping studies revealed that a nucleosome is positioned over the transcriptional initiation and the Sp1/3 binding sites. Stimulation with NaB or TSA increased histone acetylation and restriction enzyme accessibility of the ORF50 promoter transcription initiation site. Chromatin immunoprecipitation assay was used to demonstrate that the ORF50 promoter is associated with several different histone deacetylase proteins (including HDAC1, 5, and 7) in latently infected cells. NaB treatment led to the rapid association of Ini1/Snf5, a component of the Swi/Snf family of chromatin remodeling proteins, with the ORF50 promoter. Ectopic expression of the CREB-binding protein (CBP) histone acetyltransferase (HAT) stimulated plasmid-based ORF50 transcription in a HAT-dependent manner, suggesting that CBP recruitment to the ORF50 promoter can be an initiating event for transcription and viral reactivation. Together, these results suggest that remodeling of a stably positioned nucleosome at the transcriptional initiation site of ORF50 is a regulatory step in the transition from latent to lytic infection.
Figures

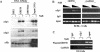
References
-
- Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. - PubMed
-
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. - PubMed
-
- Ahmad, K., and S. Henikoff. 2002. Epigenetic consequences of nucleosome dynamics. Cell 111:281-284. - PubMed
-
- Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

