Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse
- PMID: 14557630
- PMCID: PMC229353
- DOI: 10.1128/jvi.77.21.11448-11458.2003
Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse
Abstract
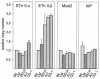
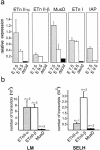
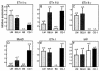
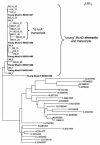
ETnII elements are mobile members of the repetitive early transposon family of mouse long terminal repeat (LTR) retroelements and have caused a number of mutations by inserting into genes. ETnII sequences lack retroviral genes, but the recent discovery of related MusD retroviral elements with regions similar to gag, pro, and pol suggests that MusD provides the proteins necessary for ETnII transposition in trans. For this study, we analyzed all ETnII elements in the draft sequence of the C57BL/6J genome and classified them into three subtypes (alpha, beta, and gamma) based on structural differences. We then used database searches and quantitative real-time PCR to determine the copy number and expression of ETnII and MusD elements in various mouse strains. In 7.5-day-old embryos of a mouse strain in which two mutations due to ETnII-beta insertions have been identified (SELH/Bc), we detected a three- to sixfold higher level of ETnII-beta and MusD transcripts than in control strains (C57BL/6J and LM/Bc). The increased ETnII transcription level can in part be attributed to a higher number of ETnII-beta elements, but 70% of the MusD transcripts appear to have been derived from one or a few MusD elements that are not detectable in C57BL/6J mice. This element belongs to a young MusD subgroup with intact open reading frames and identical LTRs, suggesting that the overexpressed element(s) in SELH/Bc mice might provide the proteins for the retrotransposition of ETnII and MusD elements. We also show that ETnII is expressed up to 30-fold more than MusD, which could explain why only ETnII, but not MusD, elements have been positively identified as new insertions.
Figures


References
-
- Baust, C., G. J. Baillie, and D. L. Mager. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13:423-428. - PubMed
-
- Dupressoir, A., A. Puech, and T. Heidmann. 1995. IAP retrotransposons in the mouse liver as reporters of ageing. Biochim. Biophys. Acta 1264:397-402. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

