Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1
- PMID: 14557648
- PMCID: PMC229369
- DOI: 10.1128/jvi.77.21.11625-11632.2003
Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1
Abstract
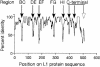
To characterize epitopes on human papillomavirus (HPV) virus-like particles (VLPs), a panel of mutated HPV-16 VLPs was created. Each mutated VLP had residues substituted from HPV-31 or HPV-52 L1 sequences to the HPV-16 L1 backbone. Mutations were created on the HPV-31 and -52 L1 proteins to determine if HPV-16 type-specific recognition could be transferred. Correct folding of the mutated proteins was verified by resistance to trypsin digestion and by binding to one or more conformation-dependent monoclonal antibodies. Several of the antibodies tested were found to bind to regions already identified as being important for HPV VLP recognition (loops DE, EF, FG, and HI). Sequences at both ends of the long FG loop (amino acids 260 to 290) were required for both H16.V5 and H16.E70 reactivity. A new antibody-binding site was discovered on the C-terminal arm of L1 between positions 427 and 445. Recognition of these residues by the H16.U4 antibody suggests that this region is surface exposed and supports a recently proposed molecular model of HPV VLPs.
Figures






References
-
- Carter, J. J., M. E. Hagensee, M. C. Taflin, S. K. Lee, L. A. Koutsky, and D. A. Galloway. 1993. HPV-1 capsids expressed in vitro detect human serum antibodies associated with foot warts. Virology 195:456-462. - PubMed
-
- Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. - PubMed
-
- Christensen, N. D., N. M. Cladel, C. A. Reed, L. R. Budgeon, M. E. Embers, D. M. Skulsky, W. L. McClements, S. W. Ludmerer, and K. U. Jansen. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324-334. - PubMed
-
- Christensen, N. D., J. Dillner, C. EkLund, J. J. Carter, G. C. Wipf, C. A. Reed, N. M. Cladel, and D. A. Galloway. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174-184. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

