Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein
- PMID: 14557649
- PMCID: PMC229354
- DOI: 10.1128/jvi.77.21.11633-11643.2003
Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein
Abstract
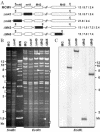
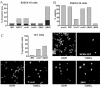
Viruses have evolved various strategies to prevent premature apoptosis of infected host cells. Some of the viral genes mediating antiapoptotic functions have been identified by their homology to cellular genes, but others are structurally unrelated to genes of known function. In this study, we used a random, unbiased approach to identify such genes in the murine cytomegalovirus genome. From a library of random transposon insertion mutants, a mutant virus that caused premature cell death was isolated. The transposon was inserted within open reading frame m41. An independently constructed m41 deletion mutant showed the same phenotype, whereas deletion mutants lacking the adjacent genes m40 and M42 did not. Apoptosis occurred in different cell types, could be blocked by caspase inhibitors, and did not require p53. Within the murine cytomegalovirus genome, m41, m40, and m39 form a small cluster of genes of unknown function. They are homologous to r41, r40, and r39 of rat cytomegalovirus, but lack sequence homology to UL41, UL40, and UL37 exon 1 (UL37x1) which are located at the corresponding positions of the human cytomegalovirus genome. Unlike UL37x1 of human cytomegalovirus, which encodes a mitochondrion-localized inhibitor of apoptosis that is essential for virus replication, m41 encodes a protein that localizes to the Golgi apparatus. The murine cytomegalovirus m41 product is the first example of a Golgi-localized protein that prevents premature apoptosis and thus extends the life span of infected cells.
Figures





References
-
- Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. - PubMed
-
- Brune, W. 2002. Random transposon mutagenesis of large DNA molecules in Escherichia coli. Methods Mol. Biol. 182:165-171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

