In silico atomic tracing by substrate-product relationships in Escherichia coli intermediary metabolism
- PMID: 14559781
- PMCID: PMC403765
- DOI: 10.1101/gr.1212003
In silico atomic tracing by substrate-product relationships in Escherichia coli intermediary metabolism
Abstract
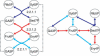
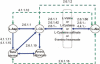
We present a software system that computationally reproduces biochemical radioisotope-tracer experiments. It consists of three main components: A mapping database of substrate-product atomic correspondents derived from known reaction formulas, a tracing engine that can compute all pathways between two given compounds by using the mapping database, and a graphical user interface. As the system can facilitate the display of all possible pathways between any two compounds and the tracing of every single carbon, nitrogen, or sulfur atom in the metabolism, it complements and bridges other metabolic databases and simulations on fixed models.
Figures
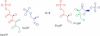

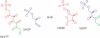
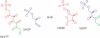

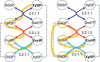
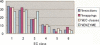
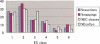
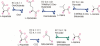



References
-
- Arita, M. 2000a. Metabolic reconstruction using shortest paths. Simulation Pract. Theory 8: 109-125.
-
- Arita, M. 2000b. Graph modeling of metabolism. J. Jpn. Soc. Artif. Intell. (JSAI) 15: 703-710.
-
- Arita, M., Asai, K., and Nishioka, T. 2000. Reconstructing metabolic pathways with new enzyme classification. In Proceedings German Conf. Bioinformatics (GCB'00), pp. 99-106. Heidelberg, Germany.
-
- Budavari, S., O'Neil, M.J., Smith, A., Heckelman, P.E., and Kinneary, J.F. eds. 1996. Merck index: An encyclopedia of chemicals, drugs, and biologicals, 12th ed. Merck, NJ.
WEB SITE REFERENCES
-
- http://us.expasy.org/enzyme; ENZYME database.
-
- http://www.brenda.uni-koeln.de; BRENDA database.
-
- http://www.chem.qmul.ac.uk/iubmb/enzyme; IUBMB enzyme nomenclature.
-
- http://www.ecocyc.org; EcoCyc database.
-
- http://www.mdli.com; MDL Information Systems.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
