Principles of cell-free genetic circuit assembly
- PMID: 14559971
- PMCID: PMC240676
- DOI: 10.1073/pnas.2135496100
Principles of cell-free genetic circuit assembly
Abstract
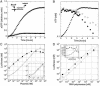
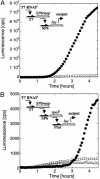
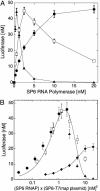
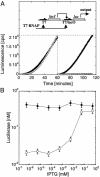
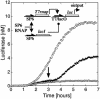
Cell-free genetic circuit elements were constructed in a transcription-translation extract. We engineered transcriptional activation and repression cascades, in which the protein product of each stage is the input required to drive or block the following stage. Although we can find regions of linear response for single stages, cascading to subsequent stages requires working in nonlinear regimes. Substantial time delays and dramatic decreases in output production are incurred with each additional stage because of a bottleneck at the translation machinery. Faster turnover of RNA message can relieve competition between genes and stabilize output against variations in input and parameters.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

