XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response
- PMID: 14559994
- PMCID: PMC207643
- DOI: 10.1128/MCB.23.21.7448-7459.2003
XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response
Abstract
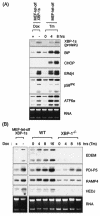
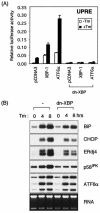
The mammalian unfolded protein response (UPR) protects the cell against the stress of misfolded proteins in the endoplasmic reticulum (ER). We have investigated here the contribution of the UPR transcription factors XBP-1, ATF6alpha, and ATF6beta to UPR target gene expression. Gene profiling of cell lines lacking these factors yielded several XBP-1-dependent UPR target genes, all of which appear to act in the ER. These included the DnaJ/Hsp40-like genes, p58(IPK), ERdj4, and HEDJ, as well as EDEM, protein disulfide isomerase-P5, and ribosome-associated membrane protein 4 (RAMP4), whereas expression of BiP was only modestly dependent on XBP-1. Surprisingly, given previous reports that enforced expression of ATF6alpha induced a subset of UPR target genes, cells deficient in ATF6alpha, ATF6beta, or both had minimal defects in upregulating UPR target genes by gene profiling analysis, suggesting the presence of compensatory mechanism(s) for ATF6 in the UPR. Since cells lacking both XBP-1 and ATF6alpha had significantly impaired induction of select UPR target genes and ERSE reporter activation, XBP-1 and ATF6alpha may serve partially redundant functions. No UPR target genes that required ATF6beta were identified, nor, in contrast to XBP-1 and ATF6alpha, did the activity of the UPRE or ERSE promoters require ATF6beta, suggesting a minor role for it during the UPR. Collectively, these results suggest that the IRE1/XBP-1 pathway is required for efficient protein folding, maturation, and degradation in the ER and imply the existence of subsets of UPR target genes as defined by their dependence on XBP-1. Further, our observations suggest the existence of additional, as-yet-unknown, key regulators of the UPR.
Figures
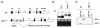



References
-
- Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. - PubMed
-
- Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. - PubMed
-
- Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
