Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2
- PMID: 14560004
- PMCID: PMC207609
- DOI: 10.1128/MCB.23.21.7566-7584.2003
Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2
Abstract
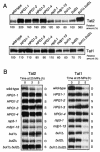
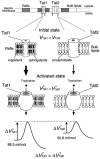
Tryptophan uptake appears to be the Achilles' heel in yeast physiology, since under a variety of seemingly diverse toxic conditions, it becomes the limiting factor for cell growth. When growing cells of Saccharomyces cerevisiae are subjected to high hydrostatic pressure, tryptophan uptake is down-regulated, leading to cell cycle arrest in the G(1) phase. Here we present evidence that the two tryptophan permeases Tat1 and Tat2 are differentially regulated by Rsp5 ubiquitin ligase in response to high hydrostatic pressure. Analysis of high-pressure growth mutants revealed that the HPG1 gene was allelic to RSP5. The HPG1 mutation or the bul1Delta bul2Delta double mutation caused a marked increase in the steady-state level of Tat2 but not of Tat1, although both permeases were degraded at high pressure in an Rsp5-dependent manner. There were marked differences in subcellular localization. Tat1 localized predominantly in the plasma membrane, whereas Tat2 was abundant in the internal membranes. Moreover, Tat1 was associated with lipid rafts, whereas Tat2 localized in bulk lipids. Surprisingly, Tat2 became associated with lipid rafts upon the occurrence of a ubiquitination defect. These results suggest that ubiquitination is an important determinant of the localization and regulation of these tryptophan permeases. Determination of the activation volume (DeltaV( not equal )) for Tat1- and Tat2-mediated tryptophan uptake (89.3 and 50.8 ml/mol, respectively) revealed that both permeases are highly sensitive to membrane perturbation and that Tat1 rather than Tat2 is likely to undergo a dramatic conformational change during tryptophan import. We suggest that hydrostatic pressure is a unique tool for elucidating the dynamics of integral membrane protein functions as well as for probing lipid microenvironments where they localize.
Figures


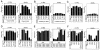





References
-
- Abe, F., and K. Horikoshi. 1998. Analysis of intracellular pH in the yeast Saccharomyces cerevisiae under elevated hydrostatic pressure: a study in baro- (piezo-) physiology. Extremophiles 2:223-228. - PubMed
-
- Abe, F., C. Kato, and K. Horikoshi. 1999. Pressure-regulated metabolism in microorganisms. Trends Microbiol. 7:447-452. - PubMed
-
- Abe, F., and K. Horikoshi. 2001. The biotechnological potential of piezophiles. Trends Biotechnol. 19:102-108. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
