Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock
- PMID: 14560008
- PMCID: PMC207607
- DOI: 10.1128/MCB.23.21.7628-7637.2003
Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock
Abstract
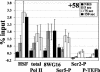
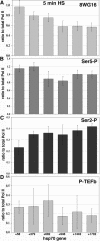
The uninduced Drosophila hsp70 gene is poised for rapid activation. Here we examine the rapid changes upon heat shock in levels and location of heat shock factor (HSF), RNA polymerase II (Pol II) and its phosphorylated forms, and the Pol II kinase P-TEFb on hsp70 in vivo by using both real-time PCR assays of chromatin immunoprecipitates and polytene chromosome immunofluorescence. These studies capture Pol II recruitment and progression along hsp70 and reveal distinct spatial and temporal patterns of serine 2 and serine 5 phosphorylation: in uninduced cells, the promoter-paused Pol II shows Ser5 but not Ser2 phosphorylation, and in induced cells the relative level of Ser2-P Pol II is lower at the promoter than at regions downstream. An early time point of heat shock activation captures unphosphorylated Pol II recruited to the promoter prior to P-TEFb, and during the first wave of transcription Pol II and the P-TEFb kinase can be seen tracking together across hsp70 with indistinguishable kinetics. Pol II distributions on several other genes with paused Pol II show a pattern of Ser5 and Ser2 phosphorylation similar to that of hsp70. These studies of factor choreography set important limits in modeling transcription regulatory mechanisms.
Figures



References
-
- Allison, L. A., M. Moyle, M. Shales, and C. J. Ingles. 1985. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 42:599-610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
