The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons
- PMID: 14563872
- PMCID: PMC219399
- DOI: 10.1128/JB.185.21.6371-6384.2003
The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons
Abstract
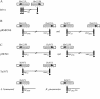
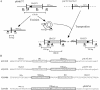
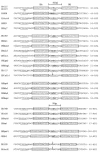
IS5075 and IS4321 are closely related (93.1% identical) members of the IS1111 family that target a specific position in the 38-bp terminal inverted repeats of Tn21 family transposons and that are inserted in only one orientation. They are 1,327 bp long and have identical ends consisting of short inverted repeats of 12 bp with an additional 7 bp (TAATGAG) or 6 bp (AATGAG) to the left of the left inverted repeats and 3 bp (AGA) or 4 bp (AGAT) to the right of the right inverted repeat. Circular forms of IS5075 and IS4321 in which the inverted repeats are separated by abutting terminal sequences (AGATAATGAG) were detected. A similar circular product was found for the related ISPa11. Transposition of IS4321 into the 38-bp target site was detected, but a flanking duplication was not generated. The precisely reconstituted target site was also identified. Over 50 members of the IS1111 family were identified. They encode related transposases, have related inverted repeats, and include related bases that lie outside these inverted repeats. In some, the flanking bases number 5 or 6 on the left and 4 or 3 on the right. Specific target sites were found for several of these insertion sequence (IS) elements. IS1111 family members therefore differ from the majority of IS elements, which are characterized by terminal inverted repeats and a target site duplication, and from members of the related IS110 family, which do not have obvious inverted repeats near their termini.
Figures



References
-
- Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
-
- Doran, T., M. Tizard, D. Millar, J. Ford, N. Sumar, M. Loughlin, and J. Hermon-Taylor. 1997. IS900 targets translation initiation signals in Mycobacterium avium subsp. paratuberculosis to facilitate expression of its hed gene. Microbiology 143:547-552. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

