Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets
- PMID: 14563874
- PMCID: PMC219383
- DOI: 10.1128/JB.185.21.6392-6399.2003
Genome-scale analysis of the uses of the Escherichia coli genome: model-driven analysis of heterogeneous data sets
Abstract
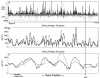
The recent availability of heterogeneous high-throughput data types has increased the need for scalable in silico methods with which to integrate data related to the processes of regulation, protein synthesis, and metabolism. A sequence-based framework for modeling transcription and translation in prokaryotes has been established and has been extended to study the expression state of the entire Escherichia coli genome. The resulting in silico analysis of the expression state highlighted three facets of gene expression in E. coli: (i) the metabolic resources required for genome expression and protein synthesis were found to be relatively invariant under the conditions tested; (ii) effective promoter strengths were estimated at the genome scale by using global mRNA abundance and half-life data, revealing genes subject to regulation under the experimental conditions tested; and (iii) large-scale genome location-dependent expression patterns with approximately 600-kb periodicity were detected in the E. coli genome based on the 49 expression data sets analyzed. These results support the notion that a structured model-driven analysis of expression data yields additional information that can be subjected to commonly used statistical analyses. The integration of heterogeneous genome-scale data (i.e., sequence, expression data, and mRNA half-life data) is readily achieved in the context of an in silico model.
Figures


References
-
- Allen, T. E., and B. O. Palsson. 2003. Sequence-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220:1-18. - PubMed
-
- Altman, R. B., and S. Raychaudhuri. 2001. Whole-genome expression analysis: challenges beyond clustering. Curr. Opin. Struct. Biol. 11:340-347. - PubMed
-
- Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. - PubMed
-
- Bentley, P. M., and J. T. E. McDonnell. 1994. Wavelet transforms: an introduction. IEE Electron. Commun. Eng. J. 6:175-186.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

