Prophage contribution to bacterial population dynamics
- PMID: 14563883
- PMCID: PMC219396
- DOI: 10.1128/JB.185.21.6467-6471.2003
Prophage contribution to bacterial population dynamics
Abstract
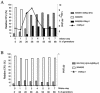
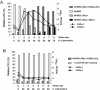
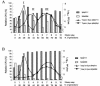
Cocultures of Salmonella strains carrying or lacking specific prophages undergo swift composition changes as a result of phage-mediated killing of sensitive bacteria and lysogenic conversion of survivors. Thus, spontaneous prophage induction in a few lysogenic cells enhances the competitive fitness of the lysogen population as a whole, setting a selection regime that forces maintenance and spread of viral DNA. This is likely to account for the profusion of prophage sequences in bacterial genomes and may contribute to the evolutionary success of certain phylogenetic lineages.
Figures

References
-
- Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. - PubMed
-
- Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

