Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition
- PMID: 14563920
- PMCID: PMC240710
- DOI: 10.1073/pnas.2034282100
Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition
Abstract
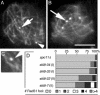
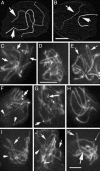
Ski8p is implicated in degradation of non-poly(A) and double-stranded RNA, and in meiotic DNA recombination. We have identified the Sordaria macrospora SKI8 gene. Ski8p is cytoplasmically localized in all vegetative and sexual cycle cells, and is nuclear localized, specifically in early-mid-meiotic prophase, in temporal correlation with Spo11p, the meiotic double-strand break (DSB) transesterase. Localizations of Ski8p and Spo11p are mutually interdependent. ski8 mutants exhibit defects in vegetative growth, entry into the sexual program, and sporulation. Diverse meiotic defects, also seen in spo11 mutants, are diagnostic of DSB absence, and they are restored by exogenous DSBs. These results suggest that Ski8p promotes meiotic DSB formation by acting directly within meiotic prophase chromosomes. Mutant phenotypes also divide meiotic homolog juxtaposition into three successive, mechanistically distinct steps; recognition, presynaptic alignment, and synapsis, which are distinguished by their differential dependence on DSBs.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

