An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3
- PMID: 14567691
- PMCID: PMC3119717
- DOI: 10.1021/bi034998x
An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3
Abstract
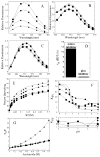
We have examined the enzymatic activity of an uncleavable procaspase-3 mutant (D9A/D28A/D175A), which contains the wild-type catalytic residues in the active site. The results are compared to those for the mature caspase-3. Although at pH 7.5 and 25 degrees C the K(m) values are similar, the catalytic efficiency (k(cat)) is approximately 130-fold lower in the zymogen. The mature caspase-3 demonstrates a maximum activity at pH 7.4, whereas the maximum activity of procaspase-3 occurs at pH 8.3. The pK(a) values of both catalytic groups, H121 and C163, are shifted to higher pH for procaspase-3. We developed limited proteolysis assays using trypsin and V8 proteases, and we show that these assays allow the examination of amino acids in three of five active site loops. In addition, we examined the fluorescence emission of the two tryptophanyl residues in the active site over the pH range of 2.5-9 as well as the response to several quenching agents. Overall, the data suggest that the major conformational change that occurs upon maturation results in formation of the loop bundle among loops L4, L2, and L2'. The pK(a) values of both catalytic groups decrease as a result of the loop movements. However, loop L3, which comprises the bulk of the substrate binding pocket, does not appear to be unraveled and solvent-exposed, even at lower pH.
Figures
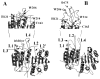





References
-
- Earnshaw WC, Martins LM, Kaufmann SH. Annu Rev Biochem. 1999;68:383–424. - PubMed
-
- Stennicke HR, Salvesen GS. Biochim Biophys Acta. 1998;1387:17–31. - PubMed
-
- Kumar S. Clin Exp Pharmacol Physiol. 1999;26:295–303. - PubMed
-
- Garcia-Calvo M, Peterson EP, Villiancourt JP, Zamboni R, Nicholson DW, Thornberry NA. Cell Death Differ. 1999;6:362–369. - PubMed
-
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Carcia-Calvo M, Hotzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. J Biol Chem. 1997;272:17907–17911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

