Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential
- PMID: 14568979
- PMCID: PMC2194228
- DOI: 10.1084/jem.20030305
Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential
Abstract
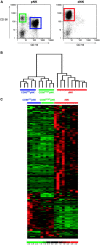
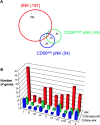
Natural killer cells constitute 50-90% of lymphocytes in human uterine decidua in early pregnancy. Here, CD56(bright) uterine decidual NK (dNK) cells were compared with the CD56(bright) and CD56(dim) peripheral NK cell subsets by microarray analysis, with verification of results by flow cytometry and RT-PCR. Among the approximately 10,000 genes studied, 278 genes showed at least a threefold change with P < or = 0.001 when comparing the dNK and peripheral NK cell subsets, most displaying increased expression in dNK cells. The largest number of these encoded surface proteins, including the unusual lectinlike receptors NKG2E and Ly-49L, several killer cell Ig-like receptors, the integrin subunits alpha(D), alpha(X), beta1, and beta5, and multiple tetraspanins (CD9, CD151, CD53, CD63, and TSPAN-5). Additionally, two secreted proteins, galectin-1 and progestagen-associated protein 14, known to have immunomodulatory functions, were selectively expressed in dNK cells.
Figures



References
-
- Billingham, R.E., L. Brent, and P.B. Medawar. 1953. Actively acquired tolerance of foreign cells. Nature. 172:603–606. - PubMed
-
- Pearson, H. 2002. Reproductive immunology: immunity's pregnant pause. Nature. 420:265–266. - PubMed
-
- Moffett-King, A. 2002. Natural killer cells and pregnancy. Nat. Rev. Immunol. 2:656–663. - PubMed
-
- Cooper, M.A., T.A. Fehniger, and M.A. Caligiuri. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640. - PubMed
-
- Faulk, W.P., and A. Temple. 1976. Distribution of beta2 microglobulin and HLA in chorionic villi of human placentae. Nature. 262:799–802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

