Genetic requirements for mycobacterial survival during infection
- PMID: 14569030
- PMCID: PMC240732
- DOI: 10.1073/pnas.2134250100
Genetic requirements for mycobacterial survival during infection
Abstract
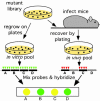
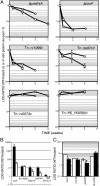
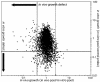
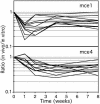
Despite the importance of tuberculosis as a public health problem, we know relatively little about the molecular mechanisms used by the causative organism, Mycobacterium tuberculosis, to persist in the host. To define these mechanisms, we have mutated virtually every nonessential gene of M. tuberculosis and determined the effect disrupting each gene on the growth rate of this pathogen during infection. A total of 194 genes that are specifically required for mycobacterial growth in vivo were identified. The behavior of these mutants provides a detailed view of the changing environment that the bacterium encounters as infection proceeds. A surprisingly large fraction of these genes are unique to mycobacteria and closely related species, indicating that many of the strategies used by this unusual group of organisms are fundamentally different from other pathogens
Figures
References
-
- Flynn, J. L. & Chan, J. (2001) Annu. Rev. Immunol. 19, 93-129. - PubMed
-
- Glickman, M. S. & Jacobs, W. R., Jr. (2001) Cell 104, 477-485. - PubMed
-
- Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2002) Mol. Microbiol. 48, 77-84. - PubMed
-
- Bardarov, S., Bardarov, S., Jr., Pavelka, M. S., Jr., Sambandamurthy, V., Larsen, M., Tufariello, J., Chan, J., Hatfull, G. & Jacobs, W. R., Jr. (2002) Microbiology 148, 3007-3017. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

