Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes
- PMID: 14572668
- PMCID: PMC7232748
- DOI: 10.1016/s0014-5793(03)01091-3
Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes
Abstract
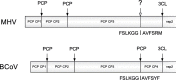
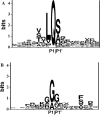
Recently, we have developed a coronavirus-specific gene-finding system, ZCURVE_CoV 1.0. In this paper, the system is further improved by taking the prediction of cleavage sites of viral proteinases in polyproteins into account. The cleavage sites of the 3C-like proteinase and papain-like proteinase are highly conserved. Based on the method of traditional positional weight matrix trained by the peptides around cleavage sites, the present method also sufficiently considers the length conservation of non-structural proteins cleaved by the 3C-like proteinase and papain-like proteinase to reduce the false positive prediction rate. The improved system, ZCURVE_CoV 2.0, has been run for each of the 24 completely sequenced coronavirus genomes in GenBank. Consequently, all the non-structural proteins in the 24 genomes are accurately predicted. Compared with known annotations, the performance of the present method is satisfactory. The software ZCURVE_CoV 2.0 is freely available at http://tubic.tju.edu.cn/sars/.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

