Identification of a 3-deoxy-D-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant
- PMID: 14573664
- PMCID: PMC219605
- DOI: 10.1128/IAI.71.11.6426-6434.2003
Identification of a 3-deoxy-D-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant
Abstract
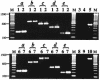
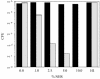
Lipooligosaccharide (LOS), a predominant surface-exposed component of the outer membrane, has been implicated as a virulence factor in the pathogenesis of Moraxella catarrhalis infections. However, the critical steps involved in the biosynthesis and assembly of M. catarrhalis LOS currently remain undefined. In this study, we used random transposon mutagenesis to identify a 3-deoxy-D-manno-octulosonic acid (KDO) biosynthetic operon in M. catarrhalis with the gene order pyrG-kdsA-eno. The lipid A-KDO molecule serves as the acceptor onto which a variety of glycosyl transferases sequentially add the core and branch oligosaccharide extensions for the LOS molecule. KdsA, the KDO-8-phosphate synthase, catalyzes the first step of KDO biosynthesis and is an essential enzyme in gram-negative enteric bacteria for maintenance of bacterial viability. We report the construction of an isogenic M. catarrhalis kdsA mutant in strain 7169 by allelic exchange. Our data indicate that an LOS molecule consisting only of lipid A and lacking KDO glycosylation is sufficient to sustain M. catarrhalis survival in vitro. In addition, comparative growth and susceptibility assays were performed to assess the sensitivity of 7169kdsA11 compared to that of the parental strain. The results of these studies demonstrate that the native LOS molecule is an important factor in maintaining the integrity of the outer membrane and suggest that LOS is a critical component involved in the ability of M. catarrhalis to resist the bactericidal activity of human sera.
Figures





Similar articles
-
Roles of 3-deoxy-D-manno-2-octulosonic acid transferase from Moraxella catarrhalis in lipooligosaccharide biosynthesis and virulence.Infect Immun. 2005 Jul;73(7):4222-30. doi: 10.1128/IAI.73.7.4222-4230.2005. Infect Immun. 2005. PMID: 15972513 Free PMC article.
-
Expression of genes kdsA and kdsB involved in 3-deoxy-D-manno-octulosonic acid metabolism and biosynthesis of enterobacterial lipopolysaccharide is growth phase regulated primarily at the transcriptional level in Escherichia coli K-12.J Bacteriol. 1995 Aug;177(15):4488-500. doi: 10.1128/jb.177.15.4488-4500.1995. J Bacteriol. 1995. PMID: 7543480 Free PMC article.
-
Genetic and biochemical characterization of an operon involved in the biosynthesis of 3-deoxy-D-manno-octulosonic acid in Pseudomonas aeruginosa.FEMS Microbiol Lett. 1999 Apr 1;173(1):27-33. doi: 10.1111/j.1574-6968.1999.tb13480.x. FEMS Microbiol Lett. 1999. PMID: 10220877
-
Conservation of the 2-keto-3-deoxymanno-octulosonic acid (Kdo) biosynthesis pathway between plants and bacteria.Carbohydr Res. 2013 Oct 18;380:70-5. doi: 10.1016/j.carres.2013.07.006. Epub 2013 Jul 20. Carbohydr Res. 2013. PMID: 23974348 Review.
-
D-galactonate metabolism in enteric bacteria: a molecular and physiological perspective.Curr Opin Microbiol. 2024 Oct;81:102524. doi: 10.1016/j.mib.2024.102524. Epub 2024 Aug 12. Curr Opin Microbiol. 2024. PMID: 39137493 Review.
Cited by
-
Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis.Infect Immun. 2010 May;78(5):2017-23. doi: 10.1128/IAI.00016-10. Epub 2010 Mar 1. Infect Immun. 2010. PMID: 20194587 Free PMC article.
-
Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation.Infect Immun. 2004 Nov;72(11):6262-70. doi: 10.1128/IAI.72.11.6262-6270.2004. Infect Immun. 2004. PMID: 15501752 Free PMC article.
-
Use of the chinchilla model for nasopharyngeal colonization to study gene expression by Moraxella catarrhalis.Infect Immun. 2012 Mar;80(3):982-95. doi: 10.1128/IAI.05918-11. Epub 2011 Dec 19. Infect Immun. 2012. PMID: 22184412 Free PMC article.
-
Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin.Infect Immun. 2006 Mar;74(3):1588-96. doi: 10.1128/IAI.74.3.1588-1596.2006. Infect Immun. 2006. PMID: 16495530 Free PMC article.
-
Modular arrangement of allelic variants explains the divergence in Moraxella catarrhalis UspA protein function.Infect Immun. 2008 Nov;76(11):5330-40. doi: 10.1128/IAI.00573-08. Epub 2008 Aug 4. Infect Immun. 2008. PMID: 18678659 Free PMC article.
References
-
- Bandak, S. I., M. R. Turnak, B. S. Allen, L. D. Bolzon, D. A. Preston, S. K. Bouchillon, and D. J. Hoban. 2001. Antibiotic susceptibilities among recent clinical isolates of Haemophilus influenzae and Moraxella catarrhalis from fifteen countries. Eur. J. Clin. Microbiol. Infect. Dis. 20:55-60. - PubMed
-
- Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46:360-371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

