A single nucleotide primer extension assay to detect the APC I1307K gene variant
- PMID: 14573780
- PMCID: PMC1907340
- DOI: 10.1016/S1525-1578(10)60477-1
A single nucleotide primer extension assay to detect the APC I1307K gene variant
Abstract
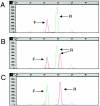
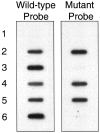
Adenomatous polyposis coli (APC) is a tumor suppressor gene important in colorectal tumorigenesis. A genetic variant of APC, I1307K, results from a T-to-A transversion at nucleotide 3920 which converts the wild-type sequence to a homopolymer tract (A(8)). The I1307K alteration is not itself oncogenic, but creates a hypermutable region (A(8)) that is prone to frame-shift mutations. The APC I1307K variant occurs in approximately 6% of the Ashkenazi Jewish population and is reported to approximately double an individual's risk for colorectal cancer. Here we describe a single nucleotide primer extension assay for the detection of the APC I1307K mutation. Following PCR amplification, nucleotide 3920 of the APC gene is directly sequenced using single nucleotide primer extension technology. The assay is in a multiplex format allowing simultaneous forward and reverse sequencing of the I1307K variant, which provides an internal, independent confirmation of each testing result. The assay was validated against 60 samples previously characterized by an allele-specific oligonucleotide (ASO) hybridization assay, with 100% concordance of results. Compared to the ASO assay, this single nucleotide primer extension assay requires significantly less technical time to perform, and has a greatly increased throughput capacity. The single nucleotide extension assay provides a highly sensitive and specific assay to identify individuals with the APC I1307K gene variant who may benefit from increased colorectal screening.
Figures

References
-
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ: Cancer statistics, 2003. CA Cancer J Clin 2003, 53:5-26 - PubMed
-
- Cannon-Albright LA, Skolnick MH, Bishop DT, Lee RG, Burt RW: Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med 1988, 319:533-537 - PubMed
-
- Fuchs CS, Giovannucci EL, Colditz GA, Hunter DJ, Speizer FE, Willett WC: A prospective study of family history and the risk of colorectal cancer. N Engl J Med 1994, 331:1669-1674 - PubMed
-
- Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 - PubMed
-
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW: APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359:235-237 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

