Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
- PMID: 14573872
- PMCID: PMC2366957
- DOI: 10.1110/ps.03256803
Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae
Abstract
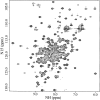
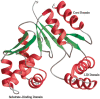
Streptococcus pneumoniae is a major human pathogen that causes high mortality and morbidity and has developed resistance to many antibiotics. We show that the gene product from SP1603, identified from S. pneumoniae TIGR4, is a CMP kinase that is essential for bacterial growth. It represents an attractive drug target for the development of a novel antibiotic to overcome the problems of drug resistance development for this organism. Here we describe the three-dimensional solution structure of the S. pneumoniae CMP kinase as determined by NMR spectroscopy. The structure consists of eight alpha-helices and two beta-sheets that fold into the classical core domain, the substrate-binding domain, and the LID domain. The three domains of the protein pack together to form a central cavity for substrate-binding and enzymatic catalysis. The S. pneumoniae CMP kinase resembles the fold of the Escherichia coli homolog. An insertion of one residue is observed at the beta-turn in the substrate-binding domain of the S. pneumoniae CMP kinase when compared with the E. coli homolog. Chemical shift perturbations caused by the binding of CMP, CDP, and ATP revealed that CMP or CDP binds to the junction between the core and substrate-binding domains, whereas ATP binds to the junction between the core and LID domains. From NMR relaxation studies, we determined that the loops in the LID domain are highly mobile. These mobile loops could aid in the closing/opening of the LID domain during enzyme catalysis.
Figures


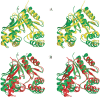
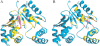

Similar articles
-
Solution structure and function of a conserved protein SP14.3 encoded by an essential Streptococcus pneumoniae gene.J Mol Biol. 2001 Aug 17;311(3):593-604. doi: 10.1006/jmbi.2001.4894. J Mol Biol. 2001. PMID: 11493012
-
Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.J Mol Biol. 2002 Feb 1;315(5):1099-110. doi: 10.1006/jmbi.2001.5286. J Mol Biol. 2002. PMID: 11827479
-
Structural and catalytic properties of CMP kinase from Bacillus subtilis: a comparative analysis with the homologous enzyme from Escherichia coli.Arch Biochem Biophys. 1997 Apr 1;340(1):144-53. doi: 10.1006/abbi.1997.9888. Arch Biochem Biophys. 1997. PMID: 9126287
-
Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.Structure. 1998 Dec 15;6(12):1517-27. doi: 10.1016/s0969-2126(98)00150-6. Structure. 1998. PMID: 9862805
-
Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity.Adv Enzymol Relat Areas Mol Biol. 1999;73:103-34, x. doi: 10.1002/9780470123195.ch4. Adv Enzymol Relat Areas Mol Biol. 1999. PMID: 10218107 Review.
Cited by
-
Structure of Staphylococcus aureus cytidine monophosphate kinase in complex with cytidine 5'-monophosphate.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006 Aug 1;62(Pt 8):710-5. doi: 10.1107/S174430910602447X. Epub 2006 Jul 24. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006. PMID: 16880539 Free PMC article.
-
A bacterial pan-genome makes gene essentiality strain-dependent and evolvable.Nat Microbiol. 2022 Oct;7(10):1580-1592. doi: 10.1038/s41564-022-01208-7. Epub 2022 Sep 12. Nat Microbiol. 2022. PMID: 36097170 Free PMC article.
-
Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6.J Bacteriol. 2007 Jan;189(1):38-51. doi: 10.1128/JB.01148-06. Epub 2006 Oct 13. J Bacteriol. 2007. PMID: 17041037 Free PMC article.
-
Molecular modeling and dynamics studies of cytidylate kinase from Mycobacterium tuberculosis H37Rv.J Mol Model. 2008 May;14(5):427-34. doi: 10.1007/s00894-008-0291-2. Epub 2008 Mar 15. J Mol Model. 2008. PMID: 18343960
-
The crystal structures of Thermus thermophilus CMP kinase complexed with a phosphoryl group acceptor and donor.PLoS One. 2020 May 29;15(5):e0233689. doi: 10.1371/journal.pone.0233689. eCollection 2020. PLoS One. 2020. PMID: 32469932 Free PMC article.
References
-
- Baquero, F. 1995. Pneumococcal resistance to β-lactam antibiotics. Microb. Drug Resist. 1 115–120. - PubMed
-
- Bertrand, T., Briozzo, P., Assairi, L., Ofiteru, A., Bucurenci, N., Munier-Lehmann, H., Golinelli-Pimpaneau, B., Barzu, O., and Gilles, A.-M. 2002. Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme. J. Mol. Biol. 315 1099–1110. - PubMed
-
- Briozzo, P., Golinelli-Pimpaneau, B., Gilles, A.-M., Gaucher, J.-F., Burlacu-Miron, S., Sakamoto, H., Janin, J., and Barzu, O. 1998. Structures of Escherichia coli CMP kinase alone and in complex with CDP: A new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure 6 1517–1527. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

