Origin and diffusion of mtDNA haplogroup X
- PMID: 14574647
- PMCID: PMC1180497
- DOI: 10.1086/379380
Origin and diffusion of mtDNA haplogroup X
Abstract
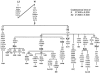
A maximum parsimony tree of 21 complete mitochondrial DNA (mtDNA) sequences belonging to haplogroup X and the survey of the haplogroup-associated polymorphisms in 13,589 mtDNAs from Eurasia and Africa revealed that haplogroup X is subdivided into two major branches, here defined as "X1" and "X2." The first is restricted to the populations of North and East Africa and the Near East, whereas X2 encompasses all X mtDNAs from Europe, western and Central Asia, Siberia, and the great majority of the Near East, as well as some North African samples. Subhaplogroup X1 diversity indicates an early coalescence time, whereas X2 has apparently undergone a more recent population expansion in Eurasia, most likely around or after the last glacial maximum. It is notable that X2 includes the two complete Native American X sequences that constitute the distinctive X2a clade, a clade that lacks close relatives in the entire Old World, including Siberia. The position of X2a in the phylogenetic tree suggests an early split from the other X2 clades, likely at the very beginning of their expansion and spread from the Near East.
Figures

References
Electronic-Database Information
-
- Estonian Biocentre, http://www.ebc.ee/EVOLUTSIOON/mtDNA-X/ (for mtDNA sequence data)
-
- Shareware Phylogenetic Network Software, http://www.fluxus-engineering.com/sharenet.htm (for Network 3111)
References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 - PubMed
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
-
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 - PubMed
-
- Bandelt H-J, Herrnstadt C, Yao Y-G, Kong Q-P, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, Howell N, Torroni A, Zhang Y-P. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet (in press) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

