The proteome of Saccharomyces cerevisiae mitochondria
- PMID: 14576278
- PMCID: PMC263752
- DOI: 10.1073/pnas.2135385100
The proteome of Saccharomyces cerevisiae mitochondria
Abstract
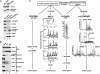
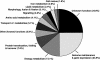
We performed a comprehensive approach to determine the proteome of Saccharomyces cerevisiae mitochondria. The proteins of highly pure yeast mitochondria were separated by several independent methods and analyzed by tandem MS. From >20 million MS spectra, 750 different proteins were identified, indicating an involvement of mitochondria in numerous cellular processes. All known components of the oxidative phosphorylation machinery, the tricarboxylic acid cycle, and the stable mitochondria-encoded proteins were found. Based on the mitochondrial proteins described in the literature so far, we calculate that the identified proteins represent approximately 90% of all mitochondrial proteins. The function of a quarter of the identified proteins is unknown. The mitochondrial proteome will provide an important database for the analysis of new mitochondrial and mitochondria-associated functions and the characterization of mitochondrial diseases.
Figures



References
-
- Schatz, G. (1995) Biochim. Biophys. Acta 1271, 123-126. - PubMed
-
- Scheffler, I. E. (1999) Mitochondria (Wiley, New York).
-
- Lill, R. & Kispal, G. (2000) Trends Biochem. Sci. 25, 352-356. - PubMed
-
- Newmeyer, D. D. & Ferguson-Miller, S. (2003) Cell 112, 481-490. - PubMed
-
- Wallace, D. C. (1999) Science 283, 1482-1488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

