Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia
- PMID: 14576291
- PMCID: PMC280571
- DOI: 10.1105/tpc.017376
Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia
Abstract
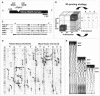
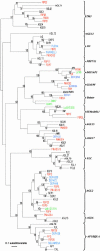
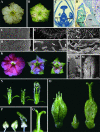
We have initiated a systematic functional analysis of the MADS box, intervening region, K domain, C domain-type MADS box gene family in petunia. The starting point for this has been a reverse-genetics approach, aiming to select for transposon insertions into any MADS box gene. We have developed and applied a family signature insertion screening protocol that is highly suited for this purpose, resulting in the isolation of 32 insertion mutants in 20 different MADS box genes. In addition, we identified three more MADS box gene insertion mutants using a candidate-gene approach. The defined insertion lines provide a sound foundation for a systematic functional analysis of the MADS box gene family in petunia. Here, we focus on the analysis of Floral Binding Protein2 (FBP2) and FBP5 genes that encode the E-function, which in Arabidopsis has been shown to be required for B and C floral organ identity functions. fbp2 mutants display sepaloid petals and ectopic inflorescences originating from the third floral whorl, whereas fbp5 mutants appear as wild type. In fbp2 fbp5 double mutants, reversion of floral organs to leaf-like organs is increased further. Strikingly, ovules are replaced by leaf-like structures in the carpel, indicating that in addition to the B- and C-functions, the D-function, which specifies ovule development, requires E-function activity. Finally, we compare our data with results obtained using cosuppression approaches and conclude that the latter might be less suited for assigning functions to individual members of the MADS box gene family.
Figures
References
-
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. - PMC - PubMed
-
- Angenent, G.C., Franken, J., Busscher, M., Colombo, L., and van Tunen, A.J. (1993). Petal and stamen formation in petunia is regulated by the homeotic gene FBP1. Plant J. 4, 101–112. - PubMed
-
- Angenent, G.C., Franken, J., Busscher, M., Weiss, D., and van Tunen, A.J. (1994). Co-suppression of the petunia homeotic gene FBP2 affects the identity of the generative meristem. Plant J. 5, 33–44. - PubMed
-
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

