Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR
- PMID: 14576300
- PMCID: PMC275455
- DOI: 10.1093/nar/gkg813
Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR
Abstract
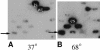
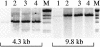
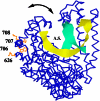
Although the thermophilic bacterium Thermus aquaticus grows optimally at 70 degrees C and cannot grow at moderate temperatures, its DNA polymerase I has significant activity at 20-37 degrees C. This activity is a bane to some PCRs, since it catalyzes non-specific priming. We report mutations of Klentaq (an N-terminal deletion variant) DNA polymerase that have markedly reduced activity at 37 degrees C yet retain apparently normal activity at 68 degrees C and resistance at 95 degrees C. The first four of these mutations are clustered on the outside surface of the enzyme, nowhere near the active site, but at the hinge point of a domain that has been proposed to move at each cycle of nucleotide incorporation. We show that the novel cold-sensitive mutants can provide a hot start for PCR and exhibit slightly improved fidelity.
Figures







Similar articles
-
Investigations on the thermostability and function of truncated Thermus aquaticus DNA polymerase fragments.Protein Eng. 1997 Nov;10(11):1281-8. doi: 10.1093/protein/10.11.1281. Protein Eng. 1997. PMID: 9514116
-
A Molecular Dynamics Investigation of the Thermostability of Cold-Sensitive I707L KlenTaq1 DNA Polymerase and Its Wild-Type Counterpart.J Chem Inf Model. 2019 May 28;59(5):2423-2431. doi: 10.1021/acs.jcim.9b00022. Epub 2019 Apr 8. J Chem Inf Model. 2019. PMID: 30897332
-
A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity.J Biol Chem. 2001 Feb 16;276(7):5044-51. doi: 10.1074/jbc.M008701200. Epub 2000 Nov 7. J Biol Chem. 2001. PMID: 11069916
-
The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion.Gene. 1992 Mar 1;112(1):29-35. doi: 10.1016/0378-1119(92)90299-5. Gene. 1992. PMID: 1551596
-
[Comparative Analysis of Family A DNA-Polymerases as a Searching Tool for Enzymes with New Properties].Mol Biol (Mosk). 2023 Mar-Apr;57(2):185-196. Mol Biol (Mosk). 2023. PMID: 37000648 Review. Russian.
Cited by
-
A Single Amino Acid Change to Taq DNA Polymerase Enables Faster PCR, Reverse Transcription and Strand-Displacement.Front Bioeng Biotechnol. 2021 Jan 14;8:553474. doi: 10.3389/fbioe.2020.553474. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33520948 Free PMC article.
-
Development of a Simple Direct and Hot-Start PCR Using Escherichia coli-Expressing Taq DNA Polymerase.Int J Mol Sci. 2023 Jul 13;24(14):11405. doi: 10.3390/ijms241411405. Int J Mol Sci. 2023. PMID: 37511160 Free PMC article.
-
Covalent modification of primers improves PCR amplification specificity and yield.Biol Methods Protoc. 2017 Nov 21;2(1):bpx011. doi: 10.1093/biomethods/bpx011. eCollection 2017 Jan. Biol Methods Protoc. 2017. PMID: 32161793 Free PMC article.
-
Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering.Front Microbiol. 2014 Sep 3;5:461. doi: 10.3389/fmicb.2014.00461. eCollection 2014. Front Microbiol. 2014. PMID: 25232352 Free PMC article.
-
Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples.Nucleic Acids Res. 2009 Apr;37(5):e40. doi: 10.1093/nar/gkn1055. Epub 2009 Feb 10. Nucleic Acids Res. 2009. PMID: 19208643 Free PMC article.
References
-
- Hebert B., Bergeron,J., Potworowski,E.F. and Tijssen,P. (1993) Increased PCR sensitivity by using paraffin wax as a reaction mix overlay. Mol. Cell. Probes, 7, 249–252. - PubMed
-
- Horton R.M., Hoppe,B.L. and Conti-Tronconik,B.M. (1994) AmpliGrease: ‘hot start’ PCR using petroleum jelly. Biotechniques, 16, 42–43. - PubMed
-
- Ramanujam R., Burdick,B.A., Landegren,U.D. and Sevigny,P. (1997) Method and preparation for sequential delivery of wax-embedded, inactivated biological and chemical reagents. US patent 5,599,660.
-
- Scalice E.R., Sharkey,D.J. and Daiss,J.L. (1994) Monoclonal antibodies prepared against the DNA polymerase from Thermus aquaticus are potent inhibitors of enzyme activity. J. Immunol. Methods, 172, 147–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

