Automated identification of RNA conformational motifs: theory and application to the HM LSU 23S rRNA
- PMID: 14576313
- PMCID: PMC275477
- DOI: 10.1093/nar/gkg835
Automated identification of RNA conformational motifs: theory and application to the HM LSU 23S rRNA
Abstract
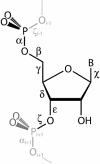
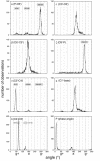
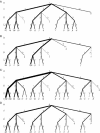
We develop novel methods for recognizing and cataloging conformational states of RNA, and for discovering statistical rules governing those states. We focus on the conformation of the large ribosomal subunit from Haloarcula marismortui. The two approaches described here involve torsion matching and binning. Torsion matching is a pattern-recognition code which finds structural repetitions. Binning is a classification technique based on distributional models of the data. In comparing the results of the two methods we have tested the hypothesis that the conformation of a very large complex RNA molecule can be described accurately by a limited number of discrete conformational states. We identify and eliminate extraneous and redundant information without losing accuracy. We conclude, as expected, that four of the torsion angles contain the overwhelming bulk of the structural information. That information is not significantly compromised by binning the continuous torsional information into a limited number of discrete values. The correspondence between torsion matching and binning is 99% (per residue). Binning, however, does have several advantages. In particular, we demonstrate that the conformation of a large complex RNA molecule can be represented by a small alphabet. In addition, the binning method lends itself to a natural graphical representation using trees.
Figures
References
-
- Cech T.R. (2001) Ribozymes, the first 20 years. Biochem. Soc. Trans, 30, 1162–1166. - PubMed
-
- Joyce G.F. (2002) The antiquity of RNA-based evolution. Nature, 418, 214–221. - PubMed
-
- Stagg S.M., Mears,J.A. and Harvey,S.C. (2003) A structural model for the assembly of the 30S subunit of the ribosome. J. Mol. Biol., 328, 49–61. - PubMed
-
- Kidd-Ljunggren K., Zuker,M., Hofacker,I.L. and Kidd,A.H. (2000) The hepatitis B virus pregenome: prediction of RNA structure and implications for the emergence of deletions. Intervirology, 43, 154–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

