Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity
- PMID: 14576318
- PMCID: PMC275449
- DOI: 10.1093/nar/gkg793
Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity
Abstract
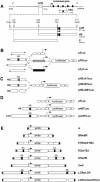
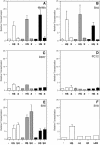
Locus control regions (LCRs) are complex high-order chromatin structures harbouring several regulatory elements, including enhancers and boundaries. We have analysed the mouse tyrosinase LCR functions, in vitro, in cell lines and, in vivo, in transgenic mice and flies. The LCR-core (2.1 kb), located at -15 kb and carrying a previously described tissue-specific DNase I hypersensitive site, operates as a transcriptional enhancer that efficiently transactivates heterologous promoters in a cell-specific orientation-independent manner. Furthermore, we have investigated the boundary activity of these sequences in transgenic animals and cells. In mice, the LCR fragment (3.7 kb) rescued a weakly expressed reference construct that displays position effects. In Drosophila, the LCR fragment and its core insulated the expression of a white minigene reporter construct from chromosomal position effects. In cells, sequences located 5' from the LCR-core displayed putative boundary activities. We have obtained genomic sequences surrounding the LCR fragment and found a LINE1 repeated element at 5'. In B16 melanoma and L929 fibroblast mouse cells, this element was found heavily methylated, supporting the existence of putative boundary elements that could prevent the spreading of condensed chromatin from the LINE1 sequences into the LCR fragment, experimentally shown to be in an open chromatin structure.
Figures


Similar articles
-
Factors binding a non-classical Cis-element prevent heterochromatin effects on locus control region activity.J Biol Chem. 2004 Apr 23;279(17):17842-9. doi: 10.1074/jbc.M401258200. Epub 2004 Feb 14. J Biol Chem. 2004. PMID: 14966120
-
Position effects are influenced by the orientation of a transgene with respect to flanking chromatin.Mol Cell Biol. 2001 Jan;21(1):298-309. doi: 10.1128/MCB.21.1.298-309.2001. Mol Cell Biol. 2001. PMID: 11113204 Free PMC article.
-
The polyoma virus enhancer cannot substitute for DNase I core hypersensitive sites 2-4 in the human beta-globin LCR.Nucleic Acids Res. 1999 Aug 1;27(15):3130-7. doi: 10.1093/nar/27.15.3130. Nucleic Acids Res. 1999. PMID: 10454609 Free PMC article.
-
Artificial chromosome transgenesis in pigmentary research.Pigment Cell Res. 2002 Aug;15(4):258-64. doi: 10.1034/j.1600-0749.2002.02030.x. Pigment Cell Res. 2002. PMID: 12100491 Review.
-
Analysis of the mouse tyrosinase promoter in vitro and in vivo.Pigment Cell Res. 1994 Oct;7(5):275-8. doi: 10.1111/j.1600-0749.1994.tb00627.x. Pigment Cell Res. 1994. PMID: 7885998 Review.
Cited by
-
Boundary sequences flanking the mouse tyrosinase locus ensure faithful pattern of gene expression.Sci Rep. 2020 Sep 23;10(1):15494. doi: 10.1038/s41598-020-72543-0. Sci Rep. 2020. PMID: 32968154 Free PMC article.
-
Regulation of the human tyrosinase gene in retinal pigment epithelium cells: the significance of transcription factor orthodenticle homeobox 2 and its polymorphic binding site.Mol Vis. 2012;18:38-54. Epub 2012 Jan 10. Mol Vis. 2012. PMID: 22259223 Free PMC article.
-
Use of transgenic animals in biotechnology: prospects and problems.Acta Naturae. 2013 Jan;5(1):33-46. Acta Naturae. 2013. PMID: 23556129 Free PMC article.
-
Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis.Nucleic Acids Res. 2015 May 26;43(10):4855-67. doi: 10.1093/nar/gkv375. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897126 Free PMC article.
-
Evolved Bmp6 enhancer alleles drive spatial shifts in gene expression during tooth development in sticklebacks.Genetics. 2021 Dec 10;219(4):iyab151. doi: 10.1093/genetics/iyab151. Genetics. 2021. PMID: 34849839 Free PMC article.
References
-
- Dillon N. and Grosveld,F. (1994) Chromatin domains as potential units of eukaryotic gene function. Curr. Opin. Genet. Dev., 4, 260–264. - PubMed
-
- Elgin S.C. (1990) Chromatin structure and gene activity. Curr. Opin. Cell. Biol., 2, 437–445. - PubMed
-
- Bell A.C., West,A.G. and Felsenfeld,G. (2001) Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science, 291, 447–450. - PubMed
-
- Laemmli U.K., Kas,E., Poljak,L. and Adachi,Y. (1992) Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev., 2, 275–285. - PubMed
-
- West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

