Intrinsic differences between authentic and cryptic 5' splice sites
- PMID: 14576320
- PMCID: PMC275472
- DOI: 10.1093/nar/gkg830
Intrinsic differences between authentic and cryptic 5' splice sites
Abstract
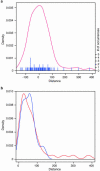
Cryptic splice sites are used only when use of a natural splice site is disrupted by mutation. To determine the features that distinguish authentic from cryptic 5' splice sites (5'ss), we systematically analyzed a set of 76 cryptic 5'ss derived from 46 human genes. These cryptic 5'ss have a similar frequency distribution in exons and introns, and are usually located close to the authentic 5'ss. Statistical analysis of the strengths of the 5'ss using the Shapiro and Senapathy matrix revealed that authentic 5'ss have significantly higher score values than cryptic 5'ss, which in turn have higher values than the mutant ones. beta-Globin provides an interesting exception to this rule, so we chose it for detailed experimental analysis in vitro. We found that the sequences of the beta-globin authentic and cryptic 5'ss, but not their surrounding context, determine the correct 5'ss choice, although their respective scores do not reflect this functional difference. Our analysis provides a statistical basis to explain the competitive advantage of authentic over cryptic 5'ss in most cases, and should facilitate the development of tools to reliably predict the effect of disease-associated 5'ss-disrupting mutations at the mRNA level.
Figures
Similar articles
-
Determinants of the inherent strength of human 5' splice sites.RNA. 2005 May;11(5):683-98. doi: 10.1261/rna.2040605. RNA. 2005. PMID: 15840817 Free PMC article.
-
Regulation of a strong F9 cryptic 5'ss by intrinsic elements and by combination of tailored U1snRNAs with antisense oligonucleotides.Hum Mol Genet. 2015 Sep 1;24(17):4809-16. doi: 10.1093/hmg/ddv205. Epub 2015 Jun 10. Hum Mol Genet. 2015. PMID: 26063760 Free PMC article.
-
Aberrant 5' splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization.Nucleic Acids Res. 2007;35(13):4250-63. doi: 10.1093/nar/gkm402. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576681 Free PMC article.
-
Pick one, but be quick: 5' splice sites and the problems of too many choices.Genes Dev. 2013 Jan 15;27(2):129-44. doi: 10.1101/gad.209759.112. Genes Dev. 2013. PMID: 23348838 Free PMC article. Review.
-
A novel role of U1 snRNP: Splice site selection from a distance.Biochim Biophys Acta Gene Regul Mech. 2019 Jun;1862(6):634-642. doi: 10.1016/j.bbagrm.2019.04.004. Epub 2019 Apr 28. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31042550 Free PMC article. Review.
Cited by
-
Targeting RNA splicing for disease therapy.Wiley Interdiscip Rev RNA. 2013 May-Jun;4(3):247-66. doi: 10.1002/wrna.1158. Epub 2013 Mar 19. Wiley Interdiscip Rev RNA. 2013. PMID: 23512601 Free PMC article. Review.
-
BAP1 missense mutation c.2054 A>T (p.E685V) completely disrupts normal splicing through creation of a novel 5' splice site in a human mesothelioma cell line.PLoS One. 2015 Apr 1;10(4):e0119224. doi: 10.1371/journal.pone.0119224. eCollection 2015. PLoS One. 2015. PMID: 25830670 Free PMC article.
-
Deep intronic mutations and human disease.Hum Genet. 2017 Sep;136(9):1093-1111. doi: 10.1007/s00439-017-1809-4. Epub 2017 May 12. Hum Genet. 2017. PMID: 28497172 Review.
-
Exonization of AluYa5 in the human ACE gene requires mutations in both 3' and 5' splice sites and is facilitated by a conserved splicing enhancer.Nucleic Acids Res. 2005 Jul 14;33(12):3897-906. doi: 10.1093/nar/gki707. Print 2005. Nucleic Acids Res. 2005. PMID: 16027113 Free PMC article.
-
Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB.BMC Mol Biol. 2007 Jan 17;8:3. doi: 10.1186/1471-2199-8-3. BMC Mol Biol. 2007. PMID: 17233885 Free PMC article.
References
-
- Horowitz D.S. and Krainer,A.R. (1994) Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet., 10, 100–106. - PubMed
-
- Zhuang Y. and Weiner,A.M. (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell, 46, 827–835. - PubMed
-
- Siliciano P.G. and Guthrie,C. (1988) 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev., 2, 1258–1267. - PubMed

