Triplex hydration: nanosecond molecular dynamics simulation of the solvated triplex formed by mixed sequences
- PMID: 14576325
- PMCID: PMC275451
- DOI: 10.1093/nar/gkg796
Triplex hydration: nanosecond molecular dynamics simulation of the solvated triplex formed by mixed sequences
Abstract
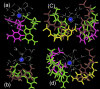
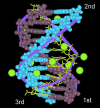
A theoretical model for the hydration pattern and motion of ions around the triple helical DNA with mixed sequences d(GACTGGTGAC)d(GTCACCAGTC)*d(GACTGGTGAC) in solution, during MD simulation, using the particle mesh Ewald sum method, is elaborated here. The AMBER 5.0 force field has been used during the simulation in solvent. The simulation studies support a dynamically stable atmosphere around the DNA triplex in solution over the entire length of the trajectory. The results have been compared with Hoogsteen triplexes and examined in the context of the observed behaviour of hydration in crystallographic data of duplexes. The dynamical organization of counterions and water molecules around the triplex formed by mixed sequences is described here. It has been observed that cations prefer to bind between two adjoining purines of the second and the third strands. The idea of localized complexes (mobile counterions in unspecific electronegative pockets around the DNA triplex with water molecules) may have important implications for understanding the specificity of the interactions of nucleic acids with proteins and other ligands.
Figures


Similar articles
-
Molecular dynamics simulation study of DNA triplex formed by mixed sequences in solution.J Biomol Struct Dyn. 2002 Aug;20(1):107-26. doi: 10.1080/07391102.2002.10506797. J Biomol Struct Dyn. 2002. PMID: 12144358
-
A 5-nanosecond molecular dynamics trajectory for B-DNA: analysis of structure, motions, and solvation.Biophys J. 1997 Nov;73(5):2313-36. doi: 10.1016/S0006-3495(97)78263-8. Biophys J. 1997. PMID: 9370428 Free PMC article.
-
Structural properties of hybrid triplex of polycation deoxyribonucleic S-methylthiourea (DNmt) strands with a complementary DNA strand, probed by nanosecond molecular dynamics.J Biomol Struct Dyn. 2000 Feb;17(4):629-43. doi: 10.1080/07391102.2000.10506554. J Biomol Struct Dyn. 2000. PMID: 10698101
-
Cations as hydrogen bond donors: a view of electrostatic interactions in DNA.Annu Rev Biophys Biomol Struct. 2003;32:27-45. doi: 10.1146/annurev.biophys.32.110601.141726. Epub 2003 Feb 14. Annu Rev Biophys Biomol Struct. 2003. PMID: 12598364 Review.
-
Potential in vivo roles of nucleic acid triple-helices.RNA Biol. 2011 May-Jun;8(3):427-39. doi: 10.4161/rna.8.3.14999. Epub 2011 May 1. RNA Biol. 2011. PMID: 21525785 Free PMC article. Review.
Cited by
-
Selective Preference of Parallel DNA Triplexes Is Due to the Disruption of Hoogsteen Hydrogen Bonds Caused by the Severe Nonisostericity between the G*GC and T*AT Triplets.PLoS One. 2016 Mar 24;11(3):e0152102. doi: 10.1371/journal.pone.0152102. eCollection 2016. PLoS One. 2016. PMID: 27010368 Free PMC article.
-
Multiscale analytic continuation approach to nanosystem simulation: applications to virus electrostatics.J Chem Phys. 2010 May 7;132(17):174112. doi: 10.1063/1.3424771. J Chem Phys. 2010. PMID: 20459161 Free PMC article.
References
-
- Westhof E. (1988) Water: an integral part of nucleic acid structure. Annu. Rev. Biophys. Biophys. Chem., 17, 125–144. - PubMed
-
- Buckin V.A. (1987) Experimental studies of DNA–water interaction. Mol. Biol., 21, 512–525.
-
- Kopka M.L., Frantini,A.V., Drew,H.R. and Dickerson,R.E. (1983) Ordered water structure around a B-DNA dodecamer. A quantitative study. J. Mol. Biol., 163, 129–146. - PubMed
-
- Saenger W., Hunter,W.N. and Kennard,O. (1986) DNA conformation is determined by economics in the hydration of phosphate groups. Nature, 324, 385–388. - PubMed
-
- Savage H. and Wlodawer,A. (1986) Determination of water structure around biomolecules using X-ray and neutron diffraction methods. Methods Enzymol., 127, 162–183. - PubMed

