Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: tailored-SELEX
- PMID: 14576330
- PMCID: PMC275487
- DOI: 10.1093/nar/gng130
Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: tailored-SELEX
Abstract
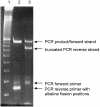
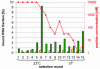
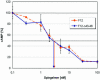
We developed an integrated method to identify aptamers with only 10 fixed nucleotides through ligation and removal of primer binding sites within the systematic evolution of ligands by exponential enrichment (SELEX) process. This Tailored-SELEX approach was validated by identifying a Spiegelmer ('mirror-image aptamer') that inhibits the action of the migraine-associated target calcitonin gene-related peptide 1 (alpha-CGRP) with an IC50 of 3 nM at 37 degrees C in cell culture. Aptamers are oligonucleotide ligands that can be generated to bind to targets with high affinity and specificity. Stabilized aptamers and Spiegelmers have shown activity in vivo and may be used as therapeutics. Aptamers are isolated by in vitro selection from combinatorial nucleic acid libraries that are composed of a central randomized region and additional fixed primer binding sites with approximately 30-40 nt. The identified sequences are usually not short enough for efficient chemical Spiegelmer synthesis, post-SELEX stabilization of aptamers and economical production. If the terminal primer binding sites are part of the target recognizing domain, truncation of aptamers has proven difficult and laborious. Tailored-SELEX results in short sequences that can be tested more rapidly in biological systems. Currently, our identified CGRP binding Spiegelmer serves as a lead compound for in vivo studies.
Figures




Similar articles
-
In vitro selection using a dual RNA library that allows primerless selection.Nucleic Acids Res. 2006 Jul 19;34(12):e86. doi: 10.1093/nar/gkl463. Nucleic Acids Res. 2006. PMID: 16855281 Free PMC article.
-
Primer-free aptamer selection using a random DNA library.J Vis Exp. 2010 Jul 26;(41):2039. doi: 10.3791/2039. J Vis Exp. 2010. PMID: 20689511 Free PMC article.
-
Capillary electrophoresis-systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: target binding dominated by 2'-O,4'-C-methylene-bridged/locked nucleic acid primer.Anal Chem. 2013 May 21;85(10):4961-7. doi: 10.1021/ac400058z. Epub 2013 May 10. Anal Chem. 2013. PMID: 23662585
-
Chemically modified nucleic acid aptamers for in vitro selections: evolving evolution.J Biotechnol. 2000 Mar;74(1):27-38. doi: 10.1016/s1389-0352(99)00002-1. J Biotechnol. 2000. PMID: 10943570 Review.
-
RNA aptamers: from basic science towards therapy.Handb Exp Pharmacol. 2006;(173):305-26. doi: 10.1007/3-540-27262-3_15. Handb Exp Pharmacol. 2006. PMID: 16594622 Review.
Cited by
-
Aptamer-iRNAs as Therapeutics for Cancer Treatment.Pharmaceuticals (Basel). 2018 Oct 18;11(4):108. doi: 10.3390/ph11040108. Pharmaceuticals (Basel). 2018. PMID: 30340426 Free PMC article. Review.
-
An L-RNA-based aquaretic agent that inhibits vasopressin in vivo.Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):5173-8. doi: 10.1073/pnas.0509663103. Epub 2006 Mar 17. Proc Natl Acad Sci U S A. 2006. PMID: 16547136 Free PMC article.
-
A mathematical analysis of SELEX.Comput Biol Chem. 2007 Feb;31(1):11-35. doi: 10.1016/j.compbiolchem.2006.10.002. Epub 2007 Jan 10. Comput Biol Chem. 2007. PMID: 17218151 Free PMC article.
-
In vitro and ex vivo selection procedures for identifying potentially therapeutic DNA and RNA molecules.Molecules. 2010 Jun 28;15(7):4610-38. doi: 10.3390/molecules15074610. Molecules. 2010. PMID: 20657381 Free PMC article. Review.
-
Updates on Aptamer Research.Int J Mol Sci. 2019 May 21;20(10):2511. doi: 10.3390/ijms20102511. Int J Mol Sci. 2019. PMID: 31117311 Free PMC article. Review.
References
-
- Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. - PubMed
-
- Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. - PubMed
-
- Ellington A.D. and Szostak,J.W. (1992) Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature, 355, 850–852. - PubMed
-
- Gold L., Polisky,B., Uhlenbeck,O. and Yarus,M. (1995) Diversity of oligonucleotide functions. Annu. Rev. Biochem., 64, 763–797. - PubMed
-
- Conrad R., Keranen,L.M., Ellington,A.D. and Newton,A.C. (1994) Isozyme-specific inhibition of protein kinase C by RNA aptamers. J. Biol. Chem., 269, 32051–32054. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

