Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease
- PMID: 14580212
- PMCID: PMC1450104
- DOI: 10.1021/bi034446e
Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease
Abstract
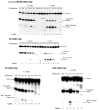
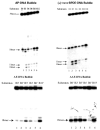
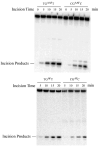
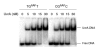
DNA damage recognition of nucleotide excision repair (NER) in Escherichia coli is achieved by at least two steps. In the first step, a helical distortion is recognized, which leads to a strand opening at the lesion site. The second step involves the recognition of the type of chemical modification in the single-stranded region of DNA during the processing of the lesions by UvrABC. In the current work, by comparing the efficiencies of UvrABC incision of several types of different DNA adducts, we show that the size and position of the strand opening are dependent on the type of DNA adducts. Optimal incision efficiency for the C8-guanine adducts of 2-aminofluorene (AF) and N-acetyl-2-aminofluorene (AAF) was observed in a bubble of three mismatched nucleotides, whereas the same for C8-guanine adduct of 1-nitropyrene and N(2)-guanine adducts of benzo[a]pyrene diol epoxide (BPDE) was noted in a bubble of six mismatched nucleotides. This suggests that the size of the aromatic ring system of the adduct might influence the extent and number of bases associated with the opened strand region catalyzed by UvrABC. We also showed that the incision efficiency of the AF or AAF adduct was affected by the neighboring DNA sequence context, which, in turn, was the result of differential binding of UvrA to the substrates. The sequence context effect on both incision and binding disappeared when a bubble structure of three bases was introduced at the adduct site. We therefore propose that these effects relate to the initial step of damage recognition of DNA structural distortion. The structure-function relationships in the recognition of the DNA lesions, based on our results, have been discussed.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

