Prediction of reduction potential changes in rubredoxin: a molecular mechanics approach
- PMID: 14581187
- PMCID: PMC1303563
- DOI: 10.1016/S0006-3495(03)74705-5
Prediction of reduction potential changes in rubredoxin: a molecular mechanics approach
Abstract
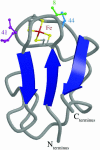
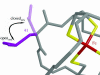
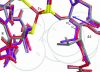
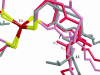
Predicting the effects of mutation on the reduction potential of proteins is crucial in understanding how reduction potentials are modulated by the protein environment. Previously, we proposed that an alanine vs. a valine at residue 44 leads to a 50-mV difference in reduction potential found in homologous rubredoxins because of a shift in the polar backbone relative to the iron site due to the different side-chain sizes. Here, the aim is to determine the effects of mutations to glycine, isoleucine, and leucine at residue 44 on the structure and reduction potential of rubredoxin, and if the effects are proportional to side-chain size. Crystal structure analysis, molecular mechanics simulations, and experimental reduction potentials of wild-type and mutant Clostridium pasteurianum rubredoxin, along with sequence analysis of homologous rubredoxins, indicate that the backbone position relative to the redox site as well as solvent penetration near the redox site are both structural determinants of the reduction potential, although not proportionally to side-chain size. Thus, protein interactions are too complex to be predicted by simple relationships, indicating the utility of molecular mechanics methods in understanding them.
Figures

References
-
- Bau, R., D. C. Rees, J. Kurtz, M. Donald, R. A. Scott, H. Huang, M. W. W. Adams, and M. K. Eidsness. 1998. Crystal structure of rubredoxin from Pyrococcus furiosus at 0.95 Å resolution, and the structures ofN-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability. J. Biol. Inorg. Chem. 3:484–493.
-
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187–217.
-
- Eidsness, M. K., A. E. Burden, K. A. Richie, D. M. J. Kurtz, R. A. Scott, E. T. Smith, T. Ichiye, B. Beard, T. Min, and C. Kang. 1999. Modulation of the redox potential of the Fe(SCys)4 site in rubredoxin by the orientation of a peptide dipole. Biochemistry. 38:14803–14809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

