Resistance of human hepatitis delta virus RNAs to dicer activity
- PMID: 14581527
- PMCID: PMC254274
- DOI: 10.1128/jvi.77.22.11910-11917.2003
Resistance of human hepatitis delta virus RNAs to dicer activity
Abstract
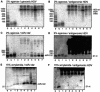
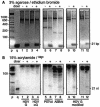
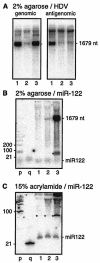
The endonuclease dicer cleaves RNAs that are 100% double stranded and certain RNAs with extensive but <100% pairing to release approximately 21-nucleotide (nt) fragments. Circular 1,679-nt genomic and antigenomic RNAs of human hepatitis delta virus (HDV) can fold into a rod-like structure with 74% pairing. However, during HDV replication in hepatocytes of human, woodchuck, and mouse origin, no approximately 21-nt RNAs were detected. Likewise, in vitro, purified recombinant dicer gave <0.2% cleavage of unit-length HDV RNAs. Similarly, rod-like RNAs of potato spindle tuber viroid (PSTVd) and avocado sunblotch viroid (ASBVd) were only 0.5% cleaved. Furthermore, when a 66-nt hairpin RNA with 79% pairing, the putative precursor to miR-122, which is an abundant liver micro-RNA, replaced one end of HDV genomic RNA, it was poorly cleaved, both in vivo and in vitro. In contrast, this 66-nt hairpin, in the absence of appended HDV sequences, was >80% cleaved in vitro. Other 66-nt hairpins derived from one end of genomic HDV, PSTVd, or ASBVd RNAs were also cleaved. Apparently, for unit-length RNAs of HDV, PSTVd, and ASBVd, it is the extended structure with <100% base pairing that confers significant resistance to dicer action.
Figures



References
-
- Asselbergs, F. A. M., and P. Grand. 1993. A two-plasmid system for transient expression of cDNAs in primate cells. Anal. Biochem. 209:327-331. - PubMed
-
- Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. - PubMed
-
- Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 195:356-372. - PubMed
-
- Cattaneo, R. 1994. Biased (adenosine to inosine) hypermutation in animal virus genomes. Curr. Opin. Genet. Dev. 4:895-900. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

