Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane
- PMID: 14581556
- PMCID: PMC254284
- DOI: 10.1128/jvi.77.22.12193-12202.2003
Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane
Abstract
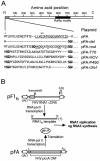
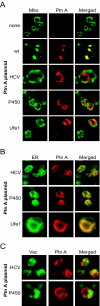
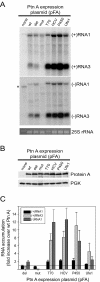
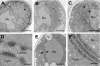
Positive-strand RNA virus replication complexes are universally associated with intracellular membranes, although different viruses use membranes derived from diverse and sometimes multiple organelles. We investigated whether unique intracellular membranes are required for viral RNA replication complex formation and function in yeast by retargeting protein A, the Flock House virus (FHV) RNA-dependent RNA polymerase. Protein A, the only viral protein required for FHV RNA replication, targets and anchors replication complexes to outer mitochondrial membranes in part via an N-proximal sequence that contains a transmembrane domain. We replaced the FHV protein A mitochondrial outer membrane-targeting sequence with the N-terminal endoplasmic reticulum (ER)-targeting sequence from the yeast NADP cytochrome P450 oxidoreductase or inverted C-terminal ER-targeting sequences from the hepatitis C virus NS5B polymerase or the yeast t-SNARE Ufe1p. Confocal immunofluorescence microscopy confirmed that protein A chimeras retargeted to the ER. FHV subgenomic and genomic RNA accumulation in yeast expressing ER-targeted protein A increased 2- to 13-fold over that in yeast expressing wild-type protein A, despite similar protein A levels. Density gradient flotation assays demonstrated that ER-targeted protein A remained membrane associated, and in vitro RNA-dependent RNA polymerase assays demonstrated an eightfold increase in the in vitro RNA synthesis activity of the ER-targeted FHV RNA replication complexes. Electron microscopy showed a change in the intracellular membrane alterations from a clustered mitochondrial distribution with wild-type protein A to the formation of perinuclear layers with ER-targeted protein A. We conclude that specific intracellular membranes are not required for FHV RNA replication complex formation and function.
Figures


References
-
- Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corp., New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

