Performance of high-throughput DNA quantification methods
- PMID: 14583097
- PMCID: PMC280658
- DOI: 10.1186/1472-6750-3-20
Performance of high-throughput DNA quantification methods
Abstract
Background: The accuracy and precision of estimates of DNA concentration are critical factors for efficient use of DNA samples in high-throughput genotype and sequence analyses. We evaluated the performance of spectrophotometric (OD) DNA quantification, and compared it to two fluorometric quantification methods, the PicoGreen assay (PG), and a novel real-time quantitative genomic PCR assay (QG) specific to a region at the human BRCA1 locus. Twenty-Two lymphoblastoid cell line DNA samples with an initial concentration of approximately 350 ng/uL were diluted to 20 ng/uL. DNA concentration was estimated by OD and further diluted to 5 ng/uL. The concentrations of multiple aliquots of the final dilution were measured by the OD, QG and PG methods. The effects of manual and robotic laboratory sample handling procedures on the estimates of DNA concentration were assessed using variance components analyses.
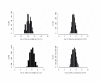
Results: The OD method was the DNA quantification method most concordant with the reference sample among the three methods evaluated. A large fraction of the total variance for all three methods (36.0-95.7%) was explained by sample-to-sample variation, whereas the amount of variance attributable to sample handling was small (0.8-17.5%). Residual error (3.2-59.4%), corresponding to un-modelled factors, contributed a greater extent to the total variation than the sample handling procedures.
Conclusion: The application of a specific DNA quantification method to a particular molecular genetic laboratory protocol must take into account the accuracy and precision of the specific method, as well as the requirements of the experimental workflow with respect to sample volumes and throughput. While OD was the most concordant and precise DNA quantification method in this study, the information provided by the quantitative PCR assay regarding the suitability of DNA samples for PCR may be an essential factor for some protocols, despite the decreased concordance and precision of this method.
Figures

References
-
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–17. - PubMed
-
- Duewer DL, Kline MC, Redman JW, Newall PJ, Reeder DJ. NIST mixed stain studies #1 and #2: interlaboratory comparison of DNA quantification practice and short tandem repeat multiplex performance with multiple-source samples. J Forensic Sci. 2001;46:1199–1210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

