A knowledge discovery object model API for Java
- PMID: 14583100
- PMCID: PMC272928
- DOI: 10.1186/1471-2105-4-51
A knowledge discovery object model API for Java
Abstract
Background: Biological data resources have become heterogeneous and derive from multiple sources. This introduces challenges in the management and utilization of this data in software development. Although efforts are underway to create a standard format for the transmission and storage of biological data, this objective has yet to be fully realized.
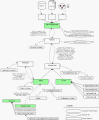
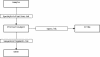
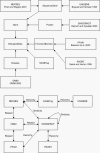
Results: This work describes an application programming interface (API) that provides a framework for developing an effective biological knowledge ontology for Java-based software projects. The API provides a robust framework for the data acquisition and management needs of an ontology implementation. In addition, the API contains classes to assist in creating GUIs to represent this data visually.
Conclusions: The Knowledge Discovery Object Model (KDOM) API is particularly useful for medium to large applications, or for a number of smaller software projects with common characteristics or objectives. KDOM can be coupled effectively with other biologically relevant APIs and classes. Source code, libraries, documentation and examples are available at http://www.bcgsc.ca/bioinfo/software.
Figures







References
-
- Uschold M, Gruninger M. Ontologies: Principles, methods and applications. Knowledge Engineering Review. 1996;11
-
- Jazayeri M. Component programming – a fresh look at software components. In: Schafer W, Botella P, editor. In Proceedings of the 5th European Software Engineering Conference: September 25–28 1995; Sitges, Spain. 1995. pp. 457–478.
-
- Sun Microsystems Java Development Kit 1.4.0. 901 San Antonio Road, Palo Alto, CA 94303. 2002. http://www.java.sun.com/j2se/1.4
-
- Stevens R, Goble CA, Bechhofer S. Ontology-based Knowledge Representation for Bioinformatics. Brief Bioinform. 2000;1:398–414. - PubMed
-
- Wiechert W, Joksch B, Wittig R, Hartbrich A, Honer T, Mollney M. Object-oriented programming for the biosciences. Bioinformatics. 1995;11:517–534. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

