Visualizing the distribution and transport of mRNAs in living cells
- PMID: 14583593
- PMCID: PMC263795
- DOI: 10.1073/pnas.2233244100
Visualizing the distribution and transport of mRNAs in living cells
Abstract
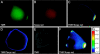
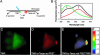
We have visualized the movements of native mRNAs in living cells. Using nuclease-resistant molecular beacons, we imaged the transport and localization of oskar mRNA in Drosophila melanogaster oocytes. When the localization pattern was altered by genetic manipulation of the mRNA's 3' untranslated region, or by chemical perturbation of the intracellular tubulin network, the distribution of the fluorescence signals changed accordingly. We tracked the migration of oskar mRNA in real time, from the nurse cells where it is produced to the posterior cortex of the oocyte where it is localized. Our observations reveal the presence of a transient, and heretofore elusive, stage in the transport of oskar mRNA. Direct visualization of specific mRNAs in living cells with molecular beacons will accelerate studies of intracellular RNA trafficking and localization, just as the use of green fluorescent protein has stimulated the study of specific proteins in vivo.
Figures

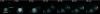
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

