Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data
- PMID: 14583755
- PMCID: PMC2394420
- DOI: 10.1038/sj.bjc.6601326
Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data
Abstract
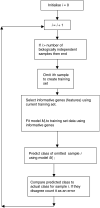
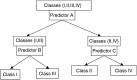
DNA microarrays are a potentially powerful technology for improving diagnostic classification, treatment selection and therapeutics development. There are, however, many potential pitfalls in the use of microarrays that result in false leads and erroneous conclusions. This paper provides a review of the key features to be observed in developing diagnostic and prognostic classification systems based on gene expression profiling and some of the pitfalls to be aware of in reading reports of microarray-based studies.
Figures
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrich JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511 - PubMed
-
- Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z (2000) Tissue classification with gene expression profiles. J Comput Biol 7: 559–584 - PubMed
-
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, VSondak Hayward N, Trent J (2000) Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406: 536–540 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

