Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha
- PMID: 14585956
- PMCID: PMC262360
- DOI: 10.1128/MCB.23.22.7947-7956.2003
Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha
Abstract
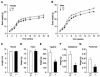
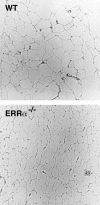
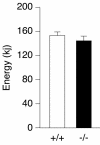
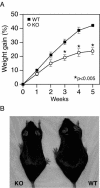
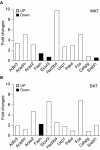
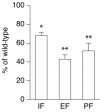
The estrogen-related receptor alpha (ERRalpha) is an orphan member of the superfamily of nuclear hormone receptors expressed in tissues that preferentially metabolize fatty acids. Despite the molecular characterization of ERRalpha and identification of target genes, determination of its physiological function has been hampered by the lack of a natural ligand. To further understand the in vivo function of ERRalpha, we generated and analyzed Estrra-null (ERRalpha-/-) mutant mice. Here we show that ERRalpha-/- mice are viable, fertile and display no gross anatomical alterations, with the exception of reduced body weight and peripheral fat deposits. No significant changes in food consumption and energy expenditure or serum biochemistry parameters were observed in the mutant animals. However, the mutant animals are resistant to a high-fat diet-induced obesity. Importantly, DNA microarray analysis of gene expression in adipose tissue demonstrates altered regulation of several enzymes involved in lipid, eicosanoid, and steroid synthesis, suggesting that the loss of ERRalpha might interfere with other nuclear receptor signaling pathways. In addition, the microarray study shows alteration in the expression of genes regulating adipogenesis as well as energy metabolism. In agreement with these findings, metabolic studies showed reduced lipogenesis in adipose tissues. This study suggests that ERRalpha functions as a metabolic regulator and that the ERRalpha-/- mice provide a novel model for the investigation of metabolic regulation by nuclear receptors.
Figures

Similar articles
-
Estrogen-related receptor alpha (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine.J Biol Chem. 2004 Dec 10;279(50):52052-8. doi: 10.1074/jbc.M410337200. Epub 2004 Oct 4. J Biol Chem. 2004. PMID: 15466464
-
Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle.Mol Cell Biol. 2004 Oct;24(20):9079-91. doi: 10.1128/MCB.24.20.9079-9091.2004. Mol Cell Biol. 2004. PMID: 15456881 Free PMC article.
-
Divergent Role of Estrogen-Related Receptor α in Lipid- and Fasting-Induced Hepatic Steatosis in Mice.Endocrinology. 2018 May 1;159(5):2153-2164. doi: 10.1210/en.2018-00115. Endocrinology. 2018. PMID: 29635284
-
ERRalpha: a metabolic function for the oldest orphan.Trends Endocrinol Metab. 2008 Oct;19(8):269-76. doi: 10.1016/j.tem.2008.07.005. Epub 2008 Sep 6. Trends Endocrinol Metab. 2008. PMID: 18778951 Free PMC article. Review.
-
The estrogen-related receptor alpha: the oldest, yet an energetic orphan with robust biological functions.J Recept Signal Transduct Res. 2010 Aug;30(4):193-205. doi: 10.3109/10799893.2010.487493. J Recept Signal Transduct Res. 2010. PMID: 20497091 Review.
Cited by
-
Loss of estrogen-related receptor alpha disrupts ventral-striatal synaptic function in female mice.Neuroscience. 2016 Aug 4;329:66-73. doi: 10.1016/j.neuroscience.2016.04.054. Epub 2016 May 4. Neuroscience. 2016. PMID: 27155145 Free PMC article.
-
Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis.PLoS One. 2010 Jul 22;5(7):e11707. doi: 10.1371/journal.pone.0011707. PLoS One. 2010. PMID: 20661474 Free PMC article.
-
Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle.Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6570-5. doi: 10.1073/pnas.0401401101. Epub 2004 Apr 20. Proc Natl Acad Sci U S A. 2004. PMID: 15100410 Free PMC article.
-
Estrogen-related receptor alpha in select host functions and cancer: new frontiers.Mol Cell Biochem. 2022 May;477(5):1349-1359. doi: 10.1007/s11010-022-04380-w. Epub 2022 Feb 9. Mol Cell Biochem. 2022. PMID: 35138514 Review.
-
Estrogen-related receptor alpha induces epithelial-mesenchymal transition through cancer-stromal interactions in endometrial cancer.Sci Rep. 2019 Apr 30;9(1):6697. doi: 10.1038/s41598-019-43261-z. Sci Rep. 2019. PMID: 31040369 Free PMC article.
References
-
- Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. - PubMed
-
- Bonnelye, E., J. M. Vanacker, T. Dittmar, A. Begue, X. Desbiens, D. T. Denhardt, J. E. Aubin, V. Laudet, and B. Fournier. 1997. The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol. Endocrinol. 11:905-916. - PubMed
-
- Chawla, A., J. J. Repa, R. M. Evans, and D. J. Mangelsdorf. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866-1870. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
