Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms
- PMID: 14585961
- PMCID: PMC262342
- DOI: 10.1128/MCB.23.22.7992-8007.2003
Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms
Abstract
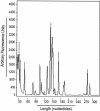
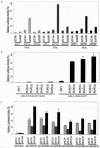
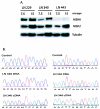
Defects in human DNA mismatch repair have been reported to underlie a variety of hereditary and sporadic cancer cases. We characterized the structure of the MSH6 promoter region to examine the mechanisms of transcriptional regulation of the MSH6 gene. The 5'-flanking region of the MSH6 gene was found to contain seven functional Sp1 transcription factor binding sites that each bind Sp1 and Sp3 and contribute to promoter activity. Transcription did not appear to require a TATA box and resulted in multiple start sites, including two major start sites and at least nine minor start sites. Three common polymorphisms were identified in the promoter region (-557 T-->G, -448 G-->A, and -159 C-->T): the latter two were always associated, and each of these functionally inactivated a different Sp1 site. The polymorphic allele -448 A -159 T was demonstrated to be a common Caucasian polymorphism found in 16% of Caucasians and resulted in a five-Sp1-site promoter that had 50% less promoter activity and was more sensitive to inactivation by DNA methylation than the more common seven Sp1 site promoter allele, which was only partially inactivated by DNA methylation. In cell lines, this five-Sp1-site polymorphism resulted in reduced MSH6 expression at both the mRNA and protein level. An additional 2% of Caucasians contained another polymorphism, -210 C-->T, which inactivated a single Sp1 site that also contributes to promoter activity.
Figures





References
-
- Akiyama, Y., H. Nagasaki, T. Nakajima, H. Sakai, T. Nomizu, and Y. Yuasa. 2001. Infrequent frameshift mutations in the simple repeat sequences of hMLH3 in hereditary nonpolyposis colorectal cancers. Jpn. J. Clin. Oncol. 31:61-64. - PubMed
-
- Andrew, S. D., P. J. Delhanty, L. M. Mulligan, and B. G. Robinson. 2000. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene 256:283-291. - PubMed
-
- Aslam, F., L. Palumbo, L. H. Augenlicht, and A. Velcich. 2001. The Sp family of transcription factors in the regulation of the human and mouse MUC2 gene promoters. Cancer Res. 61:570-576. - PubMed
-
- Bearzatto, A., M. Szadkowski, P. Macpherson, J. Jiricny, and P. Karran. 2000. Epigenetic regulation of the MGMT and hMSH6 DNA repair genes in cells resistant to methylating agents. Cancer Res. 60:3262-3270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
