Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation
- PMID: 14585976
- PMCID: PMC262398
- DOI: 10.1128/MCB.23.22.8172-8188.2003
Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation
Abstract
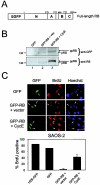
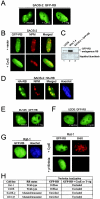
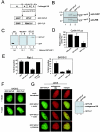
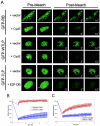
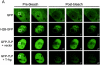
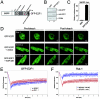
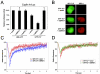
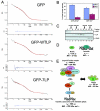
The retinoblastoma tumor suppressor, RB, assembles multiprotein complexes to mediate cell cycle inhibition. Although many RB binding partners have been suggested to underlie these functions, the validity of these interactions on the behavior of RB complexes in living cells has not been investigated. Here, we studied the dynamic behavior of RB by using green fluorescent protein-RB fusion proteins. Although these proteins were universally nuclear, phosphorylation or oncoprotein binding mediated their active exclusion from the nucleolus. In vivo imaging approaches revealed that RB exists in dynamic equilibrium between a highly mobile and a slower diffusing species, and genetic lesions associated with tumorigenesis increased the fraction of RB in a highly mobile state. The RB complexes dictating cell cycle arrest were surprisingly dynamic and harbored a relatively short residence time on chromatin. In contrast, this rapid exchange was attenuated in cells that are hypersensitive to RB, suggesting that responsiveness may inversely correlate with mobility. The stability of RB dynamics within the cell was additionally modified by the presence and function of critical corepressors. Last, the RB-assembled complexes present in living cells were primarily associated with E2F binding sites in chromatin. In contrast to RB, E2F1 consistently maintained a stable association with E2F sites regardless of cell type. Together, these results elucidate the kinetic framework of RB tumor suppressor action in transcriptional repression and cell cycle regulation.
Figures

References
-
- Angus, S. P., A. F. Fribourg, M. P. Markey, S. L. Williams, H. F. Horn, J. DeGregori, T. F. Kowalik, K. Fukasawa, and E. S. Knudsen. 2002. Active RB elicits late G1/S inhibition. Exp. Cell Res. 276:201-213. - PubMed
-
- Angus, S. P., L. J. Wheeler, S. A. Ranmal, X. Zhang, M. P. Markey, C. K. Mathews, and E. S. Knudsen. 2002. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J. Biol. Chem. 277:44376-44384. - PubMed
-
- Chew, Y. P., M. Ellis, S. Wilkie, and S. Mittnacht. 1998. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene 17:2177-2186. - PubMed
-
- Dahiya, A., S. Wong, S. Gonzalo, M. Gavin, and D. C. Dean. 2001. Linking the Rb and polycomb pathways. Mol. Cell 8:557-569. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
