Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway
- PMID: 14585983
- PMCID: PMC262401
- DOI: 10.1128/MCB.23.22.8255-8271.2003
Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway
Abstract
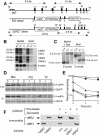
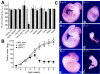
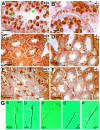
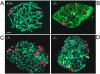
Substrates of the ubiquitin-dependent N-end rule pathway include proteins with destabilizing N-terminal residues. UBR1(-/-) mice, which lacked the pathway's ubiquitin ligase E3alpha, were viable and retained the N-end rule pathway. The present work describes the identification and analysis of mouse UBR2, a homolog of UBR1. We demonstrate that the substrate-binding properties of UBR2 are highly similar to those of UBR1, identifying UBR2 as the second E3 of the mammalian N-end rule pathway. UBR2(-/-) mouse strains were constructed, and their viability was found to be dependent on both gender and genetic background. In the strain 129 (inbred) background, the UBR2(-/-) genotype was lethal to most embryos of either gender. In the 129/B6 (mixed) background, most UBR2(-/-) females died as embryos, whereas UBR2(-/-) males were viable but infertile, owing to the postnatal degeneration of the testes. The gross architecture of UBR2(-/-) testes was normal and spermatogonia were intact as well, but UBR2(-/-) spermatocytes were arrested between leptotene/zygotene and pachytene and died through apoptosis. A conspicuous defect of UBR2(-/-) spermatocytes was the absence of intact synaptonemal complexes. We conclude that the UBR2 ubiquitin ligase and, hence, the N-end rule pathway are required for male meiosis and spermatogenesis and for an essential aspect of female embryonic development.
Figures

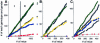

References
-
- Alfonso, P. J., and W. S. Kistler. 1993. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol. Reprod. 48:522-529. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.). 2002. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
-
- Baarends, W. M., E. Wassenaar, J. W. Hoogerbrugge, G. van Cappelen, H. P. Roest, J. Vreeburg, M. Ooms, J. H. J. Hoeijmakers, and J. A. Grootegoed. 2003. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiosis prophase. Mol. Cell. Biol. 23:1151-1162. - PMC - PubMed
-
- Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. - PubMed
-
- Buonomo, S. B. C., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103:387-398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
