Identifying inhibitors of the SARS coronavirus proteinase
- PMID: 14592491
- PMCID: PMC7119134
- DOI: 10.1016/j.bmcl.2003.08.066
Identifying inhibitors of the SARS coronavirus proteinase
Abstract
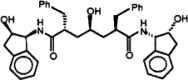
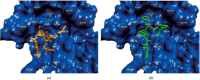
The Severe Acute Respiratory Syndrome (SARS) is a serious respiratory illness that has recently been reported in parts of Asia and Canada. In this study, we use molecular dynamics (MD) simulations and docking techniques to screen 29 approved and experimental drugs against the theoretical model of the SARS CoV proteinase as well as the experimental structure of the transmissible gastroenteritis virus (TGEV) proteinase. Our predictions indicate that existing HIV-1 protease inhibitors, L-700,417 for instance, have high binding affinities and may provide good starting points for designing SARS CoV proteinase inhibitors.
Figures
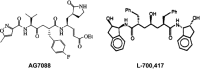
References
-
- Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. Science. 2003;300:1399. - PubMed
-
- Anand K, Ziebuhr J, Wadhwani P, Mesters J.R, Hilgenfeld R. Science. 2003;300:1763. - PubMed
-
- Clarke, T. Nature (Science Update): http://www.nature.com/nsu/030512/030512-11.html.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

