A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei
- PMID: 14592982
- PMCID: PMC275410
- DOI: 10.1093/emboj/cdg553
A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei
Abstract
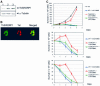
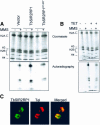
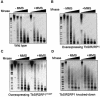
SIR2-like proteins have been implicated in a wide range of cellular events including chromosome silencing, chromosome segregation, DNA recombination and the determination of life span. We report here the molecular and functional characterization of a SIR2-related protein from the protozoan parasite Trypanosoma brucei, which we termed TbSIR2RP1. This protein is a chromosome-associated NAD-dependent enzyme which, in contrast to other known proteins of this family, catalyses both ADP-ribosylation and deacetylation of histones, particulary H2A and H2B. Under- or overexpression of TbSIR2RP1 decreased or increased, respectively, cellular resistance to DNA damage. Treatment of trypanosomal nuclei with a DNA alkylating agent resulted in a significant increase in the level of histone ADP-ribosylation and a concomitant increase in chromatin sensitivity to micrococcal nuclease. Both of these responses correlated with the level of TbSIR2RP1 expression. We propose that histone modification by TbSIR2RP1 is involved in DNA repair.
Figures




References
-
- Adamietz P. and Rudolph,A. (1984) ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem., 259, 6841–6846. - PubMed
-
- Afshar G. and Murnane,J.P. (1999) Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene, 234, 161–168. - PubMed
-
- Bell S.D., Botting,C.H., Wardleworth,B.N., Jackson,S.P. and White,M.F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science, 296, 148–151. - PubMed
-
- Bohm L., Schneeweiss,F.A., Sharan,R.N. and Feinendegen,L.E. (1997) Influence of histone acetylation on the modification of cytoplasmic and nuclear proteins by ADP-ribosylation in response to free radicals. Biochim. Biophys Acta, 1334, 149–154. - PubMed
-
- Borra M.T., O’Neill,F.J., Jackson,M.D., Marshall,B., Verdin,E., Foltz,K.R. and Denu,J.M. (2002) Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem., 277, 12632–12641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

