Exploring the functional complexity of cellular proteins by protein knockout
- PMID: 14593203
- PMCID: PMC283557
- DOI: 10.1073/pnas.2233012100
Exploring the functional complexity of cellular proteins by protein knockout
Abstract
Comprehensive dissection of protein functions entails more complicated manipulations than simply eliminating the protein of interest. Established knockdown technologies, such as RNA interference, antisense oligodeoxynucleotides, or ribozymes, are limited for specific applications such as modulating protein levels or specific targeting of a posttranslationally modified subpopulation. Here we show that the engineered Skp1, Cullin 1, and F-box-containing betaTrCP substrate receptor ubiquitin-proteolytic system, designated protein knockout, could achieve not only total elimination but also rapid and systematic reduction of a given cellular protein. Stable expression of a single engineered betaTrCP demonstrated simultaneous and sustained degradation of the entire retinoblastoma family proteins. Furthermore, the engineered betaTrCP was capable of selecting hypo- but not hyperphosphorylated forms of retinoblastoma for degradation. The engineered betaTrCP has been extensively modified to increase its specificity in substrate selection. This optimized protein-knockout system offers a powerful and versatile proteomic tool to dissect diverse functional properties of cellular proteins in somatic cells.
Figures



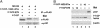

References
-
- Zhou, P., Bogacki, R., McReynolds, L. & Howley, P. M. (2000) Mol. Cell 6, 751–756. - PubMed
-
- Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. - PubMed
-
- Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., Andersen, J. S., Mann, M., Mercurio, F. & Ben-Neriah, Y. (1998) Nature 396, 590–594. - PubMed
-
- Tan, P., Fuchs, S. Y., Chen, A., Wu, K., Gomez, C., Ronai, Z. & Pan, Z. Q. (1999) Mol. Cell 3, 527–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

