Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli
- PMID: 14594828
- PMCID: PMC262112
- DOI: 10.1128/JB.185.22.6556-6561.2003
Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli
Abstract
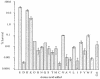
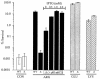
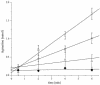
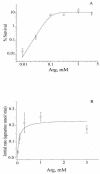
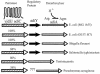
The process of arginine-dependent extreme acid resistance (XAR) is one of several decarboxylase-antiporter systems that protects Escherichia coli and possibly other enteric bacteria from exposure to the strong acid environment of the stomach. Arginine-dependent acid resistance depends on an intracellular proton-utilizing arginine alpha-decarboxylase and a membrane transport protein necessary for delivering arginine to and removing agmatine, its decarboxylation product, from the cytoplasm. The arginine system afforded significant protection to wild-type E. coli cells in our acid shock experiments. The gene coding for the transport protein is identified here as a putative membrane protein of unknown function, YjdE, which we now name adiC. Strains from which this gene is deleted fail to mount arginine-dependent XAR, and they cannot perform coupled transport of arginine and agmatine. Homologues of this gene are found in other bacteria in close proximity to homologues of the arginine decarboxylase in a gene arrangement pattern similar to that in E coli. Evidence for a lysine-dependent XAR system in E. coli is also presented. The protection by lysine, however, is milder than that by arginine.
Figures
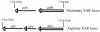

References
-
- Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. - PubMed
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Boeker, E. A., E. H. Fischer, and E. E. Snell. 1971. Arginine decarboxylase from Escherichia coli. IV. Structure of the pyridoxal phosphate binding site. J. Biol. Chem. 246:6776-6781. - PubMed
-
- Castanie-Cornet, M.-P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

